The Quick Python Book
Part 4
Working with data
In this part, you get some practice in using Python, and in particular, using it to work with data. Handling data is one of Python’s strengths. I start with basic file handling; then I move through reading from and writing to flat files, working with more structured formats such as JSON and Excel, using databases, and using Python to explore data.
These chapters are more project oriented than the rest of the book and are intended to give you the opportunity to get hands-on experience in using Python to handle data. The chapters and projects in this part can be done in any order or combination that suits your needs.
20 Basic file wrangling
This chapter covers
- Moving and renaming files
- Compressing and encrypting files
- Selectively deleting files
This chapter deals with the basic operations you can use when you have an everincreasing collection of files to manage. Those files might be log files, or they might be from a regular data feed, but whatever their source, you can’t simply discard them immediately. How do you save them, manage them, and ultimately dispose of them according to a plan but without manual intervention?
20.1 The problem: The never-ending flow of data files
Many systems generate a continuous series of data files. These files might be the log files from an e-commerce server or a regular process; they might be a nightly feed of product information from a server; they might be automated feeds of items for online advertising or historical data of stock trades; or they might come from a thousand other sources. They’re often flat text files, uncompressed, with raw data that’s either an input or a byproduct of other processes. In spite of their humble nature, however, the data they contain has some potential value, so the files can’t be discarded at the end of the day—which means that every day, their numbers grow. Over time, files accumulate until dealing with them manually becomes unworkable, and the amount of storage they consume becomes unacceptable.
20.2 Scenario: The product feed from hell
A typical situation I’ve encountered is a daily feed of product data. This data might be coming in from a supplier or going out for online marketing, but the basic aspects are the same.
Consider the example of a product feed coming from a supplier. The feed file comes in once a day, with one row for each item that the business supplies. Each row has fields for the supplier’s stock-keeping unit (SKU) number; a brief description of the item; the item’s cost, height, length, and width; the item’s status (in stock or back-ordered, say); and probably several other things, depending on the business.
In addition to this basic info file, you might well be getting others, possibly of related products, more detailed item attributes, or something else. In that case, you end up with several files with the same filenames arriving every day and landing in the same directory for processing.
Now assume that you get three related files every day: item_info.txt, item_ attributes.txt, and related_items.txt. These three files come in every day and get processed. If processing were the only requirement, you wouldn’t have to worry much; you could just let each day’s set of files replace the last and be done with it. But what if you can’t throw the data away? You may want to keep the raw data in case there’s a question about the accuracy of the process, and you need to refer to past files. Or you may want to track the changes in the data over time. Whatever the reason, the need to keep the files means that you need to do some processing.
The simplest thing you might do is mark the files with the dates on which they were received and move them to an archive folder. That way, each new set of files can be received, processed, renamed, and moved out of the way so that the process can be repeated with no loss of data.
After a few repetitions, the directory structure might look something like the following:
working/ # Main working folder, with current files for processing
item_info.txt
item_attributes.txt
related_items.txt
archive/ # Subdirectory for archiving processed files
item_info_2024-09-15.txt
item_attributes_2024-09-15.txt
related_items_2024-09-15.txt
item_info_2024-07-16.txt
item_attributes_2024-09-16.txt
related_items_2024-09-16.txt
item_info_2024-09-17.txt
item_attributes_2024-09-17.txt
related_items_2024-09-17.txt
...Think about the steps needed to make this process happen. First, you need to rename the files so that the current date is added to the filename. To do that, you need to get the names of the files you want to rename; then you need to get the stem of the filenames without the extensions. When you have the stem, you need to add a string based on the current date, add the extension back to the end, and then actually change the filename and move it to the archive directory.
What are your options for handling the tasks I’ve identified? What modules in the standard library can you think of that will do the job? If you want, you can even stop right now and work out the code to do it. Then compare your solution with the one you develop later.
You can get the names of the files in several ways. If you’re sure that the names are always exactly the same and that there aren’t many files, you could hardcode them into your script. A safer way, however, is to use the pathlib module and a path object’s glob method, as follows:
import pathlib
cur_path = pathlib.Path(".")
FILE_PATTERN = "*.txt"
path_list = cur_path.glob(FILE_PATTERN)
print(list(path_list))[PosixPath('item_attributes.txt'), PosixPath('related_items.txt'),
PosixPath('item_info.txt')]Now you can step through the paths that match your FILE_PATTERN and apply the needed changes. Remember that you need to add the date as part of the name of each file, as well move the renamed files to the archive directory. When you use pathlib, the entire operation might look like the following listing.
Listing 20.1 File files_01.py
import datetime
import pathlib
FILE_PATTERN = "*.txt" # <-- Set the pattern to match files
ARCHIVE = "archive"
def main():
date_string = datetime.date.today().strftime("%Y-%m-%d") # <-- Creates a date string based on today’s date
cur_path = pathlib.Path(".")
archive_path = cur_path.joinpath(ARCHIVE)
archive_path.mkdir(exist_ok=True) # <-- Creates an archive directory if it doesn’t exist
paths = cur_path.glob(FILE_PATTERN)
for path in paths:
new_filename = f"{path.stem}_{date_string}{path.suffix}"
new_path = archive_path.joinpath(new_filename) # <-- Creates a path from the archive path and the new filename
path.rename(new_path) # <-- Renames (and moves) the file as one step
if __name__ == '__main__':
main()The key elements in this script are finding all files that match *txt; creating a date string to add to the filenames, making sure the archive directory exists; and then moving and renaming the files. It’s worth noting here that Path objects make this operation simpler, because no special parsing is needed to separate the filename stem and suffix. This operation is also simpler than you might expect because the rename method can in effect move a file by using a path that includes the new location.
This script is a very simple one and does the job effectively in very few lines of code. In the next sections, you consider how to handle more complex requirements.
Because the preceding solution is very simple, there are likely to be many situations that it won’t handle well. What are some potential problems that might arise with the example script? How might you remedy these problems?
Consider the naming convention used for the files, which is based on the year, month, and day, in that order. What advantages do you see in that convention? What might be the disadvantages? Can you make any arguments for putting the date string somewhere else in the filename, such as the beginning or the end?
20.3 More organization
The solution to storing files described in the previous section works, but it does have some disadvantages. For one thing, as the files accumulate, managing them might become a bit more trouble, because over the course of a year, you’d have 365 sets of related files in the same directory, and you could find the related files only by inspecting their names. If the files arrive more frequently, of course, or if there are more related files in a set, the hassle would be even greater.
To mitigate this problem, you can change the way you archive the files. Instead of changing the filenames to include the dates on which they were received, you can create a separate subdirectory for each set of files and name that subdirectory after the date received. Your directory structure might look like the following:
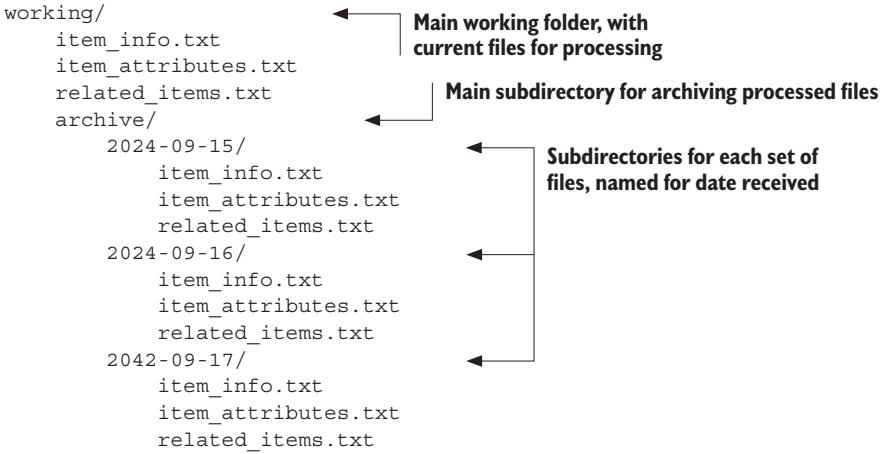
This scheme has the advantage that each set of files is grouped together. No matter how many sets of files you get or how many files you have in a set, it’s easy to find all the files of a particular set.
How would you modify the code that you developed to archive each set of files in subdirectories named according to date received? Feel free to take the time to implement the code and test it.
It turns out that archiving the files by subdirectory isn’t much more work than the first solution. The only additional step is to create the subdirectory before renaming the file. The following listing shows one way to perform this step.
Listing 20.2 File files_02.py
import datetime
import pathlib
FILE_PATTERN = "*.txt"
ARCHIVE = "archive"
def main():
date_string = datetime.date.today().strftime("%Y-%m-%d")
cur_path = pathlib.Path(".")
archive_path = cur_path.joinpath(ARCHIVE)
archive_path.mkdir(exist_ok=True) # <-- This directory needs to be created only the first time the script is run.
new_path = archive_path.joinpath(date_string)
new_path.mkdir(exist_ok=True) # <-- This directory needs to be created only once, before the files are moved into it.
paths = cur_path.glob(FILE_PATTERN)
for path in paths:
path.rename(new_path.joinpath(path.name))
if __name__ == '__main__':
main()The basic elements of this solution are the same as in the previous one but combined slightly differently. In this case, the date string is used to create a subdirectory in the archive directory and is not added to the filename. Then the file is moved to that directory but not renamed. This solution groups related files, which makes managing them as sets somewhat easier.
How might you create a script that does the same thing without using pathlib? What libraries and functions would you use?
20.4 Saving storage space: Compression and grooming
So far, you’ve been concerned mainly with managing the groups of files received. Over time, however, the data files accumulate until the amount of storage they need becomes a concern. When that happens, you have several choices. One option is to get a bigger disk. Particularly if you’re on a cloud-based platform, it may be easy and economical to adopt this strategy. Do keep in mind, however, that adding storage doesn’t really solve the problem; it merely postpones solving it.
20.4.1 Compressing files
If the space that the files are taking up is a problem, the next approach you might consider is compressing them. You have numerous ways to compress a file or set of files, but in general, these methods are similar. In this section, you consider archiving each day’s data file to a single zip file. If the files are mainly text files and are fairly large, the savings in storage achieved by compression can be impressive.
For this script, you use the same date string with a .zip extension as the name of each zip file. In listing 20.2, you created a new directory in the archive directory and then moved the files into it, which resulted in a directory structure that looks like the following:

working/ # Main working folder, where current files are processed
archive/
2024-09-15.zip # Zip files, each one containing that day's files
2024-09-16.zip # Zip files, each one containing that day's files
2024-09-17.zip # Zip files, each one containing that day's filesObviously, to use zip files, you need to change some of the steps you used previously.
Write the pseudocode for a solution that stores data files in zip files. What modules and functions or methods do you intend to use? Try coding your solution to make sure that it works.
One key addition in the new script is an import of the zipfile library and, with it, the code to create a new zipfile object in the archive directory. After that, you can use the zipfile object to write the data files to the new zip file. Finally, because you’re no longer actually moving files, you need to remove the original files from the working directory. One solution looks like the following listing.
Listing 20.3 File files_03.py
import datetime
import pathlib
import zipfile # <-- Imports zipfile library
FILE_PATTERN = "*.txt"
ARCHIVE = "archive"
def main():
date_string = datetime.date.today().strftime("%Y-%m-%d")
cur_path = pathlib.Path(".")
archive_path = cur_path.joinpath(ARCHIVE)
archive_path.mkdir(exist_ok=True)
paths = cur_path.glob(FILE_PATTERN)
zip_file_path = cur_path.joinpath(ARCHIVE, date_string + ".zip") # <-- Creates the path to the zip file in the archive directory
zip_file = zipfile.ZipFile(str(zip_file_path), "w") # <-- Opens the new zipfile object for writing
for path in paths:
zip_file.write(str(path)) # <-- Writes the current file to the zip file
path.unlink() # <-- Removes the current file from the working directory
if __name__ == '__main__':
main()This code is a bit more complex because now, instead of creating a subdirectory, we are using the date string to name and create a zipfile object. Then the contents of each file in turn are written to the zipfile object and that file is unlinked (or deleted). When the zipfile object goes out of scope at the end of the script, it is automatically closed.
20.4.2 Grooming files
Compressing data files into zip file archives can save an impressive amount of space and may be all you need. If you have a lot of files, however, or files that don’t compress much (such as JPEG image files), you may still find yourself running short of storage space. You may also find that your data doesn’t change much, making it unnecessary to keep an archived copy of every dataset in the longer term. That is, although it may be useful to keep every day’s data for the past week or month, it may not be worth the storage to keep every dataset for much longer. For data older than a few months, it may be acceptable to keep just one set of files per week or even one set per month.
The process of removing files after they reach a certain age is sometimes called grooming. Suppose that after several months of receiving a set of data files every day and archiving them in a zip file, you’re told that you should retain only one file a week of the files that are more than one month old.
The simplest grooming script removes any files that you no longer need—in this case, all but one file a week for anything older than a month old. In designing this script, it’s helpful to know the answers to two questions:
- Because you need to save one file a week, would it be much easier to simply pick the day of the week you want to save?
- How often should you do this grooming: daily, weekly, or once a month? If you decide that grooming should take place daily, it might make sense to combine the grooming with the archiving script. If, on the other hand, you need to groom only once a week or once a month, the two operations should be in separate scripts.
For this example, to keep things clear, you write a separate grooming script that can be run at any interval and that removes all the unneeded files. Further, assume that you’ve decided to keep only the files received on Tuesdays that are more than one month old. The following listing shows a sample grooming script.
Listing 20.4 File files_04.py
from datetime import datetime, timedelta
import pathlib
import zipfile
FILE_PATTERN = "*.zip"
ARCHIVE = "archive"
ARCHIVE_WEEKDAY = 1
def main():
cur_path = pathlib.Path(".")
zip_file_path = cur_path.joinpath(ARCHIVE)
paths = zip_file_path.glob(FILE_PATTERN)
current_date = datetime.today() # <-- Gets a datetime object for the current day
for path in paths:
name = path.stem # <-- Stem = filename without any extension
path_date = datetime.strptime(name, "%Y-%m-%d") # <-- strptime parses a string into a datetime object.
path_timedelta = current_date - path_date # <-- Subtracting one date from another yields a timedelta object.
if (path_timedelta > timedelta(days=30) # <-- timedelta(days = 30) creates a timedelta object of 30 days; the weekday() method returns an integer for the day of the week, with Monday = 0.
and path_date.weekday() != ARCHIVE_WEEKDAY):
path.unlink()
if __name__ == '__main__':
main()The code shows how Python’s datetime and pathlib libraries can be combined to groom files by date with only a few lines of code. Because your archive files have names derived from the dates on which they were received, you can get those file paths by using the glob method, extract the stem, and use strptime to parse the stem into a datetime object. From there, you can use datetime’s timedelta objects and the weekday() method to find a file’s age and the day of the week and then remove (unlink) the files that have passed the time limit.
Take some time to consider different grooming options. How would you modify the code in listing 20.4 to keep only one file a month? How would you change it so that files from the previous month and older were groomed to save one a week? (Note: This is not the same as older than 30 days!)
Summary
- The pathlib module can greatly simplify file operations, such as finding the root and extension, moving and renaming, and matching wildcards.
- As the number and complexity of files increases, automated archiving solutions are vital, and Python offers several easy ways to create them.
- Moving data to specific directories is as easy as renaming the files and may make managing them easier.
- You can dramatically save storage space by writing to compressed archives, such as zip files.
- As the amount of data grows, it is often useful to groom data files by deleting files that have a certain age.
21 Processing data files
This chapter covers
- Using extract-transform-load
- Reading text data files (plain text and CSV)
- Reading spreadsheet files
- Normalizing, cleaning, and sorting data
- Writing data files
Much of the data available is contained in text files. This data can range from unstructured text, such as a corpus of tweets or literary texts, to more structured data in which each row is a record and the fields are delimited by a special character, such as a comma, a tab, or a pipe (|). Text files can be huge; a dataset can be spread over tens or even hundreds of files, and the data in it can be incomplete or horribly dirty. With all the variations, it’s almost inevitable that you’ll need to read and use data from text files. This chapter gives you strategies for using Python to do exactly that.
21.1 Welcome to ETL
The need to get data out of files, parse it, turn it into a useful format, and then do something with it has been around for as long as there have been data files. In fact, there is a standard term for the process: extract-transform-load (ETL). The extraction refers to the process of reading a data source and parsing it, if necessary. The transformation can be cleaning and normalizing the data, as well as combining, breaking up, or reorganizing the records it contains. The loading refers to storing the transformed data in a new place, either a different file or a database. This chapter deals with the basics of ETL in Python, starting with text-based data files and storing the transformed data in other files. I look at more structured data files in chapter 23 and storage in databases in chapter 24.
21.2 Reading text files
The first part of ETL—the “extract” portion—involves opening a file and reading its contents. This process seems like a simple one, but even at this point there can be problems, such as the file’s size. If a file is too large to fit into memory and be manipulated, you need to structure your code to handle smaller segments of the file, possibly operating one line at a time.
21.2.1 Text encoding: ASCII, Unicode, and others
Another possible pitfall is in the encoding. This chapter deals with text files, and in fact, much of the data exchanged in the real world is in text files. But the exact nature of text can vary from application to application, from person to person, and of course from country to country.
Sometimes, “text” means something in ASCII encoding, which has 128 characters, only 95 of which are printable. The good news about ASCII encoding is that it’s the lowest common denominator of most data exchange. The bad news is that it doesn’t begin to handle the complexities of the many alphabets and writing systems of the world. Reading files using ASCII encoding is almost certain to cause trouble and throw errors on character values that it doesn’t understand, whether it’s a German ü, a Portuguese ç, or something from almost any language other than English.
These errors arise because ASCII is based on 7-bit values, whereas the bytes in a typical file are 8 bits, allowing 256 possible values as opposed to the 128 of a 7-bit value. It’s routine to use those additional values to store additional characters—anything from extra punctuation (such as the printer’s en dash and em dash) to symbols (such as the trademark, copyright, and degree symbols) to accented versions of alphabetical characters. The problem has always been that if, in reading a text file, you encounter a character in the 128 values outside the ASCII range, you have no way of knowing for sure how it was encoded. Is the character value of 214, for example, a division symbol? an Ö? or something else? Short of having the code that created the file, you have no way of knowing.
Unicode and UTF-8
One way to mitigate this confusion is Unicode. The Unicode encoding called UTF-8 accepts the basic ASCII characters without any change but also allows an almost unlimited set of other characters and symbols according to the Unicode standard. Because of its flexibility, UTF-8 was used in 98% of web pages served at the time of writing, which means that your best bet for reading text files is to assume UTF-8 encoding. If the files contain only ASCII characters, they’ll still be read correctly, but you’ll also be covered if other characters are encoded in UTF-8. The good news is that the Python 3 string data type was designed to handle Unicode by default.
Even with Unicode, there’ll be occasions when your text contains values that can’t be successfully encoded. Fortunately, the open function in Python accepts an optional errors parameter that tells it how to deal with encoding errors when reading or writing files. The default option is ‘strict’, which causes an error to be raised whenever an encoding error is encountered. Other useful options are ‘ignore’, which causes the character causing the error to be skipped; ‘replace’, which causes the character to be replaced by a marker character (often, ?); ‘backslashreplace’, which replaces the character with a backslash escape sequence; and ‘surrogateescape’, which translates the offending character to a private Unicode code point on reading and back to the original sequence of bytes on writing. Your particular use case will determine how strict you need to be in handling or resolving encoding problems.
Consider a short example of a file containing an invalid UTF-8 character and see how the different options handle that character. First, write the file, using bytes and binary mode:
open('test.txt', 'wb').write(bytes([65, 66, 67, 255, 192,193]))This code results in a file that contains “ABC” followed by three non-ASCII characters, which may be rendered differently depending on the encoding used. If you use an editor like Vim to look at the file, you might see
ABCÿÀÁNow that you have the file, try reading it with the default ‘strict’ errors option:
x = open('test.txt').read()---------------------------------------------------------------------------
UnicodeDecodeError Traceback (most recent call last)
<ipython-input-2-2cb3105c1e5f> in <cell line: 1>()
----> 1 x = open('test.txt').read()
/usr/lib/python3.10/codecs.py in decode(self, input, final)
320 # decode input (taking the buffer into account)
321 data = self.buffer + input
--> 322 (result, consumed) = self._buffer_decode(data, self.errors,
final)
323 # keep undecoded input until the next call
324 self.buffer = data[consumed:]
UnicodeDecodeError: 'utf-8' codec can't decode byte 0xff in position 3: invalid start byteThe fourth byte, which had a value of 255, isn’t a valid UTF-8 character in that position, so the ‘strict’ errors setting raises an exception. Now see how the other error options handle the same file, keeping in mind that the last three characters raise an error:
open('test.txt', errors='ignore').read()
#> 'ABC'
open('test.txt', errors='replace').read()
#> 'ABC ' # <-- Replaced error characters with chr(65533)
open('test.txt', errors='surrogateescape').read()
#> 'ABC\udcff\udcc0\udcc1'
open('test.txt', errors='backslashreplace').read()
#> 'ABC\\xff\\xc0\\xc1'If you want any problem characters to disappear, ‘ignore’ is the option to use. The ‘replace’ option only marks the place occupied by the invalid character, and the other options in different ways attempt to preserve the invalid characters without interpretation.
21.2.2 Unstructured text
Unstructured text files are the easiest sort of data to read but the hardest to extract information from. Processing unstructured text can vary enormously, depending on both the nature of the text and what you want to do with it, so any comprehensive discussion of text processing is beyond the scope of this book. A short example, however, can illustrate some of the basic topics and set the stage for a discussion of structured text data files.
One of the simplest concerns is deciding what forms a basic logical unit in the file. If you have a corpus of thousands of tweets, the text of Moby Dick, or a collection of news stories, you need to be able to break them up into cohesive units. In the case of tweets, each may fit onto a single line, and you can read and process each line of the file fairly simply.
In the case of Moby Dick or even a news story, the problem can be trickier. You may not want to treat all of a novel or news item as a single item, in many cases. But if that’s the case, you need to decide what sort of unit you do want and then come up with a strategy to divide the file accordingly. Perhaps you want to consider the text paragraph by paragraph. In that case, you need to identify how paragraphs are separated in your file and create your code accordingly. If a paragraph is the same as a line in the text file, the job is easy. Often, however, the line breaks in a text file are shorter, and you need to do a bit more work.
Now look at a couple of examples:
Call me Ishmael. Some years ago--never mind how long precisely--
having little or no money in my purse, and nothing particular
to interest me on shore, I thought I would sail about a little
and see the watery part of the world. It is a way I have
of driving off the spleen and regulating the circulation.
Whenever I find myself growing grim about the mouth;
whenever it is a damp, drizzly November in my soul; whenever I
find myself involuntarily pausing before coffin warehouses,
and bringing up the rear of every funeral I meet;
and especially whenever my hypos get such an upper hand of me,
that it requires a strong moral principle to prevent me from
deliberately stepping into the street, and methodically knocking
people's hats off--then, I account it high time to get to sea
as soon as I can. This is my substitute for pistol and ball.
With a philosophical flourish Cato throws himself upon his sword;
I quietly take to the ship. There is nothing surprising in this.
If they but knew it, almost all men in their degree, some time
or other, cherish very nearly the same feelings towards
the ocean with me.
There now is your insular city of the Manhattoes, belted round by wharves
as Indian isles by coral reefs--commerce surrounds it with her surf.
Right and left, the streets take you waterward. Its extreme downtown
is the battery, where that noble mole is washed by waves, and cooled
by breezes, which a few hours previous were out of sight of land.
Look at the crowds of water-gazers there.In the sample, which is indeed the beginning of Moby Dick, the lines are broken more or less as they might be on the page, and paragraphs are indicated by a single blank line. If you want to deal with each paragraph as a unit, you need to break the text on the blank lines. Fortunately, this task is easy if you use the string split() method. Each newline character in a string can represented by “”. Naturally, the last line of a paragraph’s text ends with a newline, and if the next line is blank, it’s immediately followed by a second newline for the blank line:
moby_text = open("moby_01.txt").read() # <-- Reads all of file as a single string
moby_paragraphs = moby_text.split("\n\n") # <-- Splits on two newlines together
print(moby_paragraphs[1])There now is your insular city of the Manhattoes, belted round by wharves
as Indian isles by coral reefs--commerce surrounds it with her surf.
Right and left, the streets take you waterward. Its extreme downtown
is the battery, where that noble mole is washed by waves, and cooled
by breezes, which a few hours previous were out of sight of land.
Look at the crowds of water-gazers there.Splitting the text into paragraphs can be done by splitting on two newlines-moby_text.split("\n\n"). Such splitting is a very simple first step in handling unstructured text, and you might also need to do more normalization of the text before processing. Suppose that you want to count the rate of occurrence of every word in a text file. If you just split the file on whitespace, you get a list of words in the file. Counting their occurrences accurately will be hard, however, because This, this, this., and this, are not the same. The way to make this code work is to normalize the text by removing the punctuation and making everything the same case before processing. For the preceding example text, the code for a normalized list of words might look like this:
moby_text = open("moby_01.txt").read() # <-- Reads all of the file as a single string
moby_paragraphs = moby_text.split("\n\n")
moby = moby_paragraphs[1].lower() # <-- Makes everything lowercase
moby = moby.replace(".", "") # <-- Removes periods
moby = moby.replace(",", "") # <-- Removes commas
moby_words = moby.split()
print(moby_words)['there', 'now', 'is', 'your', 'insular', 'city', 'of', 'the',
'manhattoes,', 'belted', 'round', 'by', 'wharves', 'as', 'indian', 'isles'
'by', 'coral', 'reefs--commerce', 'surrounds', 'it', 'with', 'her',
'surf', 'right', 'and', 'left,', 'the', 'streets', 'take', 'you',
'waterward', 'its', 'extreme', 'downtown', 'is', 'the', 'battery,',
'where', 'that', 'noble', 'mole', 'is', 'washed', 'by', 'waves,', 'and',
'cooled', 'by', 'breezes,', 'which', 'a', 'few', 'hours', 'previous',
'were', 'out', 'of', 'sight', 'of', 'land', 'look', 'at', 'the', 'crowds',
'of', 'water-gazers', 'there']Here we use the .lower string method first and then use the .replace method twice to remove two punctuation characters. Of course, as we saw back in chapter 6, it would probably be more efficient to use the string .translate method to replace all of the punctuation in one operation.
Look closely at the list of words generated. Do you see any problems with the normalization so far? What other problems do you think you might encounter in a longer section of text? How do you think you might deal with those problems?
21.2.3 Delimited flat files
Although reading unstructured text files is easy, the downside is their very lack of structure. It’s often much more useful to have some organization in the file to help with picking out individual values. The simplest way is to break the file into lines and have one element of information per line. You may have a list of the names of files to be processed, a list of people’s names that need to be printed (on name tags, say), or maybe a series of temperature readings from a remote monitor. In such cases, the data parsing is very simple: you read in the line and convert it to the right type, if necessary. Then the file is ready to use.
Most of the time, however, things aren’t quite so simple. Usually, you need to group multiple related bits of information, and you need your code to read them in together. The common way to do this is to put the related pieces of information on the same line, separated by a special character. That way, as you read each line of the file, you can use the special characters to split the file into its different fields and put the values of those fields in variables for later processing.
The following listing shows a simple example of temperature data in delimited format.
Listing 21.1 temp_data_pipes_00a.txt
State|Month Day, Year Code|Avg Daily Max Air Temperature (F)|Record Count
➥for Daily Max Air Temp (F)
Illinois|1979/01/01|17.48|994
Illinois|1979/01/02|4.64|994
Illinois|1979/01/03|11.05|994
Illinois|1979/01/04|9.51|994
Illinois|1979/05/15|68.42|994
Illinois|1979/05/16|70.29|994
Illinois|1979/05/17|75.34|994
Illinois|1979/05/18|79.13|994
Illinois|1979/05/19|74.94|994This data is pipe delimited, meaning that each field in the line is separated by the pipe (|) character—in this case, giving you four fields: the state of the observations, the date of the observations, the average high temperature, and the number of stations reporting. Other common delimiters are the tab character and the comma. The comma is perhaps the most common, but the delimiter could be any character you don’t expect to occur in the values. (More about that topic next.) Comma delimiters are so common that this format is often called comma-separated values (CSV), and files of this type often have a .csv extension as a hint of their format.
Whatever character is being used as the delimiter, if you know what character it is, you can write your own code in Python to break each line into its fields and return them as a list. In the previous case, you can use the string split() method to break a line into a list of values:
line = "Illinois|1979/01/01|17.48|994"
print(line.split("|"))
#> ['Illinois', '1979/01/01', '17.48', '994']Note that this technique is very easy to do but leaves all the values as strings, which might not be convenient for later processing.
Write the code to read a text file (assume the file is temp_data_ pipes_00a.txt, as shown in the example), split each line of the file into a list of values, and add that list to a single list of records.
What concerns or problems did you encounter in implementing this code? How might you go about converting the last three fields to the correct date, real, and int types?
21.2.4 The csv module
If you need to do much processing of delimited data files, you should become familiar with the csv module and its options. When I’ve been asked to name my favorite module in the Python standard library, more than once I’ve cited the csv module—not because it’s glamorous (it isn’t) but because it has probably saved me more work and kept me from more self-inflicted bugs over my career than any other module.
The csv module is a perfect case of Python’s “batteries included” philosophy. Although it’s perfectly possible, and in many cases not even terribly hard, to roll your own code to read delimited files, it’s even easier and much more reliable to use the Python module. The csv module has been tested and optimized, and it has features that you probably wouldn’t bother to write if you had to do it yourself but that are truly handy and time saving when available.
Look at the previous data and decide how you’d read it by using the csv module. The code to parse the data has to do two things: read each line and strip off the trailing newline character and then break up the line on the pipe character and append that list of values to a list of lines. Your solution to the exercise might look something like the following:
results = []
for line in open("temp_data_pipes_00a.txt"):
fields = line.strip().split("|")
results.append(fields)
print(results)[['Notes'],
['State',
'Month Day, Year Code',
'Avg Daily Max Air Temperature (F)',
'Record Count for Daily Max Air Temp (F)'],
['Illinois', '1979/01/01', '17.48', '994'],
['Illinois', '1979/01/02', '4.64', '994'],
['Illinois', '1979/01/03', '11.05', '994'],
['Illinois', '1979/01/04', '9.51', '994'],
['Illinois', '1979/05/15', '68.42', '994'],
['Illinois', '1979/05/16', '70.29', '994'],
['Illinois', '1979/05/17', '75.34', '994'],
['Illinois', '1979/05/18', '79.13', '994'],
['Illinois', '1979/05/19', '74.94', '994']]To do the same thing with the csv module, the code might be something like
import csv
results = [fields for fields in csv.reader(open("temp_data_pipes_00a.txt",
newline=''), delimiter="|")]
results[['State', 'Month Day, Year Code', 'Avg Daily Max Air Temperature (F)',
'Record Count for Daily Max Air Temp (F)'], ['Illinois', '1979/01/01',
'17.48', '994'], ['Illinois', '1979/01/02', '4.64', '994'], ['Illinois',
'1979/01/03', '11.05', '994'], ['Illinois', '1979/01/04', '9.51', '994'],
['Illinois', '1979/05/15', '68.42', '994'], ['Illinois', '1979/05/16',
'70.29', '994'], ['Illinois', '1979/05/17', '75.34', '994'], ['Illinois',
'1979/05/18', '79.13', '994'], ['Illinois', '1979/05/19', '74.94', '994']]In this simple case, the gain over rolling your own code doesn’t seem so great. Still, the code is two lines shorter and a bit clearer, and there’s no need to worry about stripping off newline characters. The real advantages come when you want to deal with more challenging cases.
The data in the example is real, but it’s actually been simplified and cleaned. The real data from the source is more complex. The real data has more fields, some fields are in quotes while others are not, and the first field is empty. The original is tab-delimited, but for the sake of illustration, I present it as comma delimited here.
Listing 21.2 temp_data_01.csv
"Notes","State","State Code","Month Day, Year","Month Day, Year Code",Avg
Daily Max Air Temperature (F),Record Count for Daily Max Air Temp (F),Min
Temp for Daily Max Air Temp (F),Max Temp for Daily Max Air Temp (F),Avg
Daily Max Heat Index (F),Record Count for Daily Max Heat Index (F),Min for
Daily Max Heat Index (F),Max for Daily Max Heat Index (F),Daily Max Heat
Index (F) % Coverage
,"Illinois","17","Jan 01,
1979","1979/01/01",17.48,994,6.00,30.50,Missing,0,Missing,Missing,0.00%
,"Illinois","17","Jan 02,
1979","1979/01/02",4.64,994,-6.40,15.80,Missing,0,Missing,Missing,0.00%
,"Illinois","17","Jan 03,
1979","1979/01/03",11.05,994,-0.70,24.70,Missing,0,Missing,Missing,0.00%
,"Illinois","17","Jan 04,
1979","1979/01/04",9.51,994,0.20,27.60,Missing,0,Missing,Missing,0.00%
,"Illinois","17","May 15,
1979","1979/05/15",68.42,994,61.00,75.10,Missing,0,Missing,Missing,0.00%
,"Illinois","17","May 16,
1979","1979/05/16",70.29,994,63.40,73.50,Missing,0,Missing,Missing,0.00%
,"Illinois","17","May 17, 1979","1979/05/17",75.34,994,64.00,80.50,82.60,2,82
.40,82.80,0.20%
,"Illinois","17","May 18, 1979","1979/05/18",79.13,994,75.50,82.10,81.42,349,
80.20,83.40,35.11%
,"Illinois","17","May 19, 1979","1979/05/19",74.94,994,66.90,83.10,82.87,78,8
1.60,85.20,7.85%Notice that some fields include commas. The convention in that case is to put quotes around a field to indicate that it’s not supposed to be parsed for delimiters. It’s quite common, as here, to quote only some fields, especially those in which a value might contain the delimiter character. It also happens, as here, that some fields are quoted even if they’re not likely to contain the delimiting character.
In a case like this one, your homegrown code becomes cumbersome. Now you can no longer split the line on the delimiting character; you need to be sure that you look only at delimiters that aren’t inside quoted strings. Also, you need to remove the quotes around quoted strings, which might occur in any position or not at all. With the csv module, you don’t need to change your code at all. In fact, because the comma is the default delimiter, you don’t even need to specify it:
results2 = [fields for fields in csv.reader(open("temp_data_01.csv",
newline=''))]
results2[['Notes', 'State', 'State Code', 'Month Day, Year', 'Month Day, Year
Code', 'Avg Daily Max Air Temperature (F)', 'Record Count for Daily Max Air
Temp (F)', 'Min Temp for Daily Max Air Temp (F)', 'Max Temp for Daily Max
Air Temp (F)', 'Avg Daily Max Heat Index (F)', 'Record Count for Daily Max
Heat Index (F)', 'Min for Daily Max Heat Index (F)', 'Max for Daily Max
Heat Index (F)', 'Daily Max Heat Index (F) % Coverage'], [], ['',
'Illinois', '17', 'Jan 01, 1979', '1979/01/01', '17.48', '994', '6.00',
'30.50', 'Missing', '0', 'Missing', 'Missing', '0.00%'], ['', 'Illinois',
'17', 'Jan 02, 1979', '1979/01/02', '4.64', '994', '-6.40', '15.80',
'Missing', '0', 'Missing', 'Missing', '0.00%'], ['', 'Illinois', '17', 'Jan
03, 1979', '1979/01/03', '11.05', '994', '-0.70', '24.70', 'Missing', '0',
'Missing', 'Missing', '0.00%'], ['', 'Illinois', '17', 'Jan 04, 1979',
'1979/01/04', '9.51', '994', '0.20', '27.60', 'Missing', '0', 'Missing',
'Missing', '0.00%'], ['', 'Illinois', '17', 'May 15, 1979', '1979/05/15',
'68.42', '994', '61.00', '75.10', 'Missing', '0', 'Missing', 'Missing',
'0.00%'], ['', 'Illinois', '17', 'May 16, 1979', '1979/05/16', '70.29',
'994', '63.40', '73.50', 'Missing', '0', 'Missing', 'Missing', '0.00%'],
['', 'Illinois', '17', 'May 17, 1979', '1979/05/17', '75.34', '994',
'64.00', '80.50', '82.60', '2', '82.40', '82.80', '0.20%'], ['',
'Illinois', '17', 'May 18, 1979', '1979/05/18', '79.13', '994', '75.50',
'82.10', '81.42', '349', '80.20', '83.40', '35.11%'], ['', 'Illinois',
'17', 'May 19, 1979', '1979/05/19', '74.94', '994', '66.90', '83.10',
\'82.87', '78', '81.60', '85.20', '7.85%']]Notice that the extra quotes have been removed and that any field values with commas have the commas intact inside the fields—all without any more characters in the command.
Consider how you’d approach the problems of handling quoted fields and embedded delimiter characters if you didn’t have the csv library. Which would be easier to handle: the quoting or the embedded delimiters?
21.2.5 Reading a csv file as a list of dictionaries
In the preceding examples, you got a row of data back as a list of fields. This result works fine in many cases, but sometimes it may be handy to get the rows back as dictionaries where the field name is the key. For this use case, the csv library has a DictReader, which can take a list of fields as a parameter or can read them from the first line of the data. If you want to open the data with a DictReader, the code would look like this:
results = [fields for fields in csv.DictReader(open("temp_data_01.csv",
newline=''))]
results[0]{'Notes': '',
'State': 'Illinois',
'State Code': '17',
'Month Day, Year': 'Jan 01, 1979',
'Month Day, Year Code': '1979/01/01',
'Avg Daily Max Air Temperature (F)': '17.48',
'Record Count for Daily Max Air Temp (F)': '994',
'Min Temp for Daily Max Air Temp (F)': '6.00',
'Max Temp for Daily Max Air Temp (F)': '30.50',
'Avg Daily Max Heat Index (F)': 'Missing',
'Record Count for Daily Max Heat Index (F)': '0',
'Min for Daily Max Heat Index (F)': 'Missing',
'Max for Daily Max Heat Index (F)': 'Missing',
'Daily Max Heat Index (F) % Coverage': '0.00%'}Note that, with a dictionary, the fields stay in their original order:
results[0]['State']'Illinois'If the data is particularly complex, and specific fields need to be manipulated, a DictReader can make it much easier to be sure you’re getting the right field; it also makes your code somewhat easier to understand. Conversely, if your dataset is quite large, you need to keep in mind that DictReader can take on the order of twice as long to read the same amount of data.
21.3 Excel files
The other common file format that I discuss in this chapter is the Excel file, which is the format that Microsoft Excel uses to store spreadsheets. I include Excel files here because the way you end up treating them is very similar to the way you treat delimited files. In fact, because Excel can both read and write CSV files, the quickest and easiest way to extract data from an Excel spreadsheet file often is to open it in Excel and then save it as a CSV file. This procedure doesn’t always make sense, however, particularly if you have a lot of files. In that case, even though you could theoretically automate the process of opening and saving each file in CSV format, it’s probably faster to deal with the Excel files directly.
It’s beyond the scope of this book to have an in-depth discussion of spreadsheet files, with their options for multiple sheets in the same file, macros, and various formatting options. Instead, in this section, I look at an example of reading a simple one-sheet file simply to extract the data from it.
As it happens, Python’s standard library doesn’t have a module to read or write Excel files. To read that format, you need to install an external module. Fortunately, several modules are available to do the job. For this example, you use one called openpyxl, which is available from the Python package repository. You can install it with the following command from a command line:
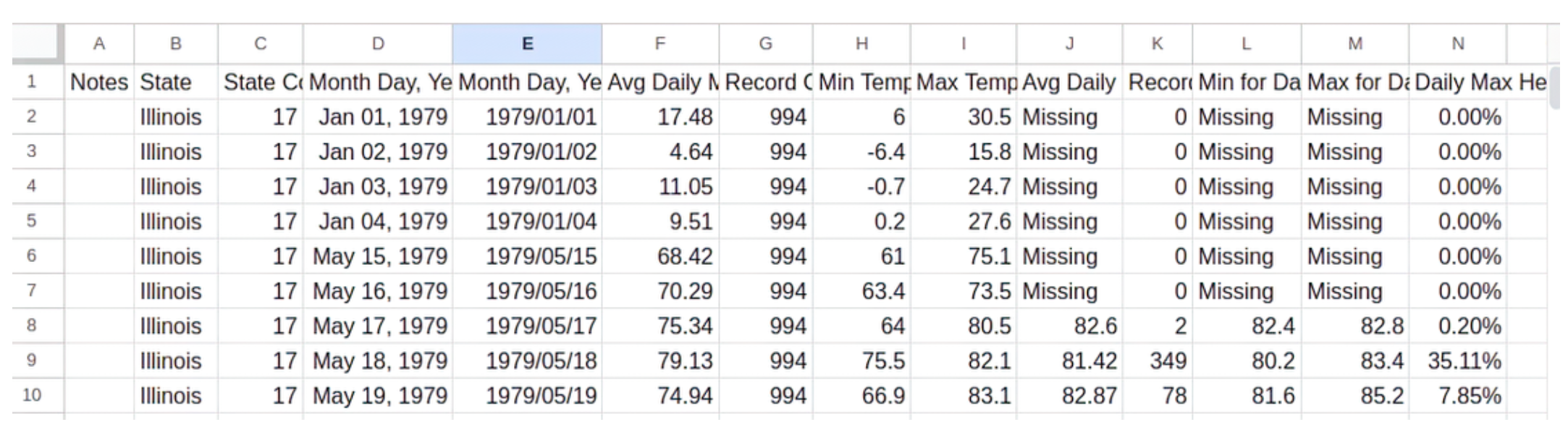
pip install openpyxlFigure 21.1 is a view of the previous temperature data, but in a spreadsheet.
Reading the file is fairly simple, but it’s still more work than CSV files require. First, you need to load the workbook; next, you need to get the specific sheet; then, you can iterate over the rows; and from there, you extract the values of the cells. Some sample code to read the spreadsheet looks like the following:
from openpyxl import load_workbook
wb = load_workbook('temp_data_01.xlsx')
results = []
ws = wb.worksheets[0]
for row in ws.iter_rows():
results.append([cell.value for cell in row])
print(results)[['Notes', 'State', 'State Code', 'Month Day, Year', 'Month Day, Year
Code', 'Avg Daily Max Air Temperature (F)', 'Record Count for Daily Max Air
Temp (F)', 'Min Temp for Daily Max Air Temp (F)', 'Max Temp for Daily Max
Air Temp (F)', 'Avg Daily Max Heat Index (F)', 'Record Count for Daily Max
Heat Index (F)', 'Min for Daily Max Heat Index (F)', 'Max for Daily Max
Heat Index (F)', 'Daily Max Heat Index (F) % Coverage'], [None, 'Illinois',
17, 'Jan 01, 1979', '1979/01/01', 17.48, 994, 6, 30.5, 'Missing', 0,
'Missing', 'Missing', '0.00%'], [None, 'Illinois', 17, 'Jan 02, 1979',
'1979/01/02', 4.64, 994, -6.4, 15.8, 'Missing', 0, 'Missing', 'Missing',
'0.00%'], [None, 'Illinois', 17, 'Jan 03, 1979', '1979/01/03', 11.05, 994,
-0.7, 24.7, 'Missing', 0, 'Missing', 'Missing', '0.00%'], [None,
'Illinois', 17, 'Jan 04, 1979', '1979/01/04', 9.51, 994, 0.2, 27.6,
'Missing', 0, 'Missing', 'Missing', '0.00%'], [None, 'Illinois', 17, 'May
15, 1979', '1979/05/15', 68.42, 994, 61, 75.1, 'Missing', 0, 'Missing',
'Missing', '0.00%'], [None, 'Illinois', 17, 'May 16, 1979', '1979/05/16',
70.29, 994, 63.4, 73.5, 'Missing', 0, 'Missing', 'Missing', '0.00%'],
[None, 'Illinois', 17, 'May 17, 1979', '1979/05/17', 75.34, 994, 64,
80.5,82.6, 2, 82.4, 82.8, '0.20%'], [None, 'Illinois', 17, 'May 18, 1979',
'1979/05/18', 79.13, 994, 75.5, 82.1, 81.42, 349, 80.2, 83.4,
'35.11%'],[None, 'Illinois', 17, 'May 19, 1979', '1979/05/19', 74.94,
994,66.9, 83.1, 82.87, 78, 81.6, 85.2, '7.85%']]This code gets you the same results as the much simpler code did for a CSV file. It’s not surprising that the code to read a spreadsheet is more complex, because spreadsheets are themselves much more complex objects. You should also be sure that you understand the way that data has been stored in the spreadsheet. If the spreadsheet contains formatting that has some significance, if labels need to be disregarded or handled differently, or if formulas and references need to be processed, you need to dig deeper into how those elements should be processed, and you need to write more complex code.
Spreadsheets also often have other possible problems. As of this writing, it’s common for spreadsheets to be limited to around a million rows. Although that limit sounds large, more and more often you’ll need to handle datasets that are larger. Also, spreadsheets sometimes automatically apply inconvenient formatting. One company I worked for had part numbers that consisted of a digit and at least one letter followed by some combination of digits and letters. It was possible to get a part number such as 1E20. Most spreadsheets automatically interpret 1E20 as scientific notation and save it as 1.00E + 20 (1 × 10 to the 20th power) while leaving 1F20 as a string. For some reason, it’s rather difficult to keep this from happening, and particularly with a large dataset, the problem won’t be detected until farther down the pipeline, if all. For these reasons, I recommend using CSV or delimited files when at all possible. Users usually can save a spreadsheet as CSV, so there’s no need to put up with the extra complexity and formatting hassles that spreadsheets involve.
21.4 Data cleaning
One common problem you’ll encounter in processing text-based data files is dirty data. By dirty, I mean that there are all sorts of surprises in the data, such as null values, values that aren’t legal for your encoding, or extra whitespace. The data may also be unsorted or in an order that makes processing difficult. The process of dealing with situations like these is called data cleaning.
21.4.1 Cleaning
In a very simple example, you might need to process a file that was exported from a spreadsheet or other financial program, and the columns dealing with money may have percentage and currency symbols (such as %, $, £, and €), as well as extra groupings that use a period or comma. Data from other sources may have other surprises that make processing tricky if they’re not caught in advance. Look again at the temperature data you saw previously. The first data line looks like this:
[None, 'Illinois', 17, 'Jan 01, 1979', '1979/01/01', 17.48, 994, 6, 30.5,
2.89, 994, -13.6, 15.8, 'Missing', 0, 'Missing', 'Missing', '0.00%']Some columns, such as ‘State’ (field 2) and ‘Notes’ (field 1), are clearly text, and you wouldn’t be likely to do much with them. There are also two date fields in different formats, and you might well want to do calculations with the dates, possibly to change the order of the data and to group rows by month or day or possibly to calculate how far apart in time two rows are.
The rest of the fields seem to be different types of numbers; the temperatures are decimals, and the record counts columns are integers. Notice, however, that the heat index temperatures have a variation: when the value for the ‘Max Temp for Daily Max Air Temp (F)’ field is below 80, the values for the heat index fields aren’t reported but instead are listed as ‘Missing’, and the record count is 0. Also note that the ‘Daily Max Heat Index (F) % Coverage’ field is expressed as a percentage of the number of temperature records that also qualify to have a heat index. Both of these things will be problematic if you want to do any math calculations on the values in those fields, because both ‘Missing’ and any number ending with % will be parsed as strings, not numbers.
Cleaning data like this can be done at different steps in the process. Quite often, I prefer to clean the data as it’s being read from the file, so I might well replace the ‘Missing’ with a None value or an empty string as the lines are being processed. You could also leave the ‘Missing’ strings in place and write your code so that no math operations are performed on a value if it is ‘Missing’.
How would you handle the fields with ‘Missing’ as possible values for math calculations? Can you write a snippet of code that averages one of those columns?
What would you do with the Daily Max Heat Index (F) % Coverage column at the end so that you could also report the average coverage? In your opinion, would the solution to this problem be at all linked to the way that the ‘Missing’ entries were handled?
21.4.2 Sorting
As I mentioned earlier, it’s often useful to have data in the text file sorted before processing. Sorting the data makes it easier to spot and handle duplicate values, and it can also help bring together related rows for quicker or easier processing. In one case, I received a 20 million–row file of attributes and values, in which arbitrary numbers of them needed to be matched with items from a master SKU list. Sorting the rows by the item ID made gathering each item’s attributes much faster. How you do the sorting depends on the size of the data file relative to your available memory and on the complexity of the sort. If all the lines of the file can fit comfortably into available memory, the easiest thing may be to read all of the lines into a list and use the list’s sort method. For example, if datafile contained the following lines:
ZZZZZZ
CCCCCC
QQQQQQ
AAAAAAthen we could sort them this way:
lines = open("datafile").readlines()
lines.sort()
print(lines)['AAAAAA\n', 'CCCCCC\n', 'QQQQQQ\n', 'ZZZZZZ\n']You could also use the sorted() function, as in sorted_lines = sorted(lines). This function preserves the order of the lines in your original list, which usually is unnecessary. The drawback to using the sorted() function is that it creates a new copy of the list. This process takes slightly longer and consumes twice as much memory, which might be a bigger concern.
If the dataset is larger than memory and the sort is very simple (just by an easily grabbed field), it may be easier to use an external utility, such as the UNIX sort command, to preprocess the data:
! sort datafile > datafile.srt
! cat datafile.srtAAAAAA
CCCCCC
QQQQQQ
ZZZZZZIn either case, sorting can be done in reverse order and can be keyed by values, not the beginning of the line. For such occasions, you need to study the documentation of the sorting tool you choose to use. A simple example in Python would be to make a sort of lines of text case insensitive. To do this, you give the sort method a key function that makes the element lowercase before making a comparison:
lines.sort(key=str.lower)This example uses a lambda function to ignore the first five characters of each string:
lines.sort(key=lambda x: x[5:])Using key functions to determine the behavior of sorts in Python is very handy, but be aware that the key function is called a lot in the process of sorting, so a complex key function could mean a real performance slowdown, particularly with a large dataset.
21.4.3 Data cleaning problems and pitfalls
It seems that there are as many types of dirty data as there are sources and use cases for that data. Your data will always have quirks that do everything from making processing less accurate to making it impossible to even load the data. As a result, I can’t provide an exhaustive list of the problems you might encounter and how to deal with them, but I can give you some general hints:
- Beware of whitespace and null characters. The problem with whitespace characters is that you can’t see them, but that doesn’t mean that they can’t cause trouble. Extra whitespace at the beginning and end of data lines, extra whitespace around individual fields, and tabs instead of spaces (or vice versa) can all make your data loading and processing more troublesome, and these problems aren’t always easily apparent. Similarly, text files with null characters (ASCII 0) may seem okay on inspection but break on loading and processing.
- Beware of punctuation. Punctuation can also be a problem. Extra commas or periods can mess up CSV files and the processing of numeric fields, and unescaped or unmatched quote characters can also confuse things.
- Break down and debug the steps. It’s easier to debug a problem if each step is separate, which means putting each operation on a separate line, being more verbose, and using more variables. But the work is worth it. For one thing, it makes any exceptions that are raised easier to understand, and it also makes debugging easier, whether with print statements, logging, or the Python debugger. It may also be helpful to save the data after each step and to cut the file size to just a few lines that cause error.
21.5 Writing data files
The last part of the ETL process may involve saving the transformed data to a database (which I discuss in chapter 22), but often it involves writing the data to files. These files may be used as input for other applications and analysis, either by people or by other applications. Usually, you have a particular file specification listing what fields of data should be included, what they should be named, what format and constraints there should be for each, and so on.
21.5.1 CSV and other delimited files
Probably the easiest thing of all is to write your data to CSV files. Because you’ve already loaded, parsed, cleaned, and transformed the data, you’re unlikely to hit any unresolved problems with the data itself. And again, using the csv module from the Python standard library makes your work much easier.
Writing delimited files with the csv module is pretty much the reverse of the read process. Again, you need to specify the delimiter that you want to use, and again, the csv module takes care of any situations in which your delimiting character is included in a field:
temperature_data = [['State', 'Month Day, Year Code', 'Avg Daily Max Air
Temperature (F)', 'Record Count for Daily Max Air Temp (F)'], ['Illinois',
'1979/01/01', '17.48', '994'], ['Illinois', '1979/01/02', '4.64', '994'],
['Illinois', '1979/01/03', '11.05', '994'], ['Illinois', '1979/01/04',
'9.51', '994'], ['Illinois', '1979/05/15', '68.42', '994'], ['Illinois',
'1979/05/16', '70.29', '994'], ['Illinois', '1979/05/17', '75.34', '994'],
['Illinois', '1979/05/18', '79.13', '994'], ['Illinois', '1979/05/19',
'74.94', '994']]
csv.writer(open("temp_data_03.csv", "w",
newline='')).writerows(temperature_data)This code results in the following file:
State,"Month Day, Year Code",Avg Daily Max Air Temperature (F),Record Coun
➥ for Daily Max Air Temp (F)
Illinois,1979/01/01,17.48,994
Illinois,1979/01/02,4.64,994
Illinois,1979/01/03,11.05,994
Illinois,1979/01/04,9.51,994
Illinois,1979/05/15,68.42,994
Illinois,1979/05/16,70.29,994
Illinois,1979/05/17,75.34,994
Illinois,1979/05/18,79.13,994
Illinois,1979/05/19,74.94,994Just as when reading from a CSV file, it’s possible to write dictionaries instead of lists if you use a DictWriter. If you do use a DictWriter, be aware of a couple of points: you must specify the field names in a list when you create the writer, and you can use the DictWriter’s writeheader method to write the header at the top of the file. So assume that you have the same data as previously but in dictionary format:
data = [{'State': 'Illinois',
'Month Day, Year Code': '1979/01/01',
'Avg Daily Max Air Temperature (F)': '17.48',
'Record Count for Daily Max Air Temp (F)': '994'}]
fields = ['State', 'Month Day, Year Code',
'Avg Daily Max Air Temperature (F)',
'Record Count for Daily Max Air Temp (F)']You can use a DictWriter object from the csv module to write each row, a dictionary, to the correct fields in the CSV file:
fields = ['State', 'Month Day, Year Code',
'Avg Daily Max Air Temperature (F)',
'Record Count for Daily Max Air Temp (F)']
dict_writer = csv.DictWriter(open("temp_data_04.csv", "w"),
fieldnames=fields)
dict_writer.writeheader()
dict_writer.writerows(data)
del dict_writer21.5.2 Writing Excel files
Writing spreadsheet files is unsurprisingly similar to reading them. You need to create a workbook, or spreadsheet file; then, you need to create a sheet or sheets; and finally, you write the data in the appropriate cells. You could create a new spreadsheet from your CSV data file like this:
from openpyxl import Workbook
data_rows = [fields for fields in csv.reader(open("temp_data_01.csv"))]
wb = Workbook()
ws = wb.active
ws.title = "temperature data"
for row in data_rows:
ws.append(row)
wb.save("temp_data_02.xlsx")It’s also possible to add formatting to cells as you write them to the spreadsheet file. For more on how to add formatting, please refer to the xlswriter documentation.
21.5.3 Packaging data files
If you have several related data files, or if your files are large, it may make sense to package them in a compressed archive. Although various archive formats are in use, the zip file remains popular and almost universally accessible to users on almost every platform. For hints on how to create zipfile packages of your data files, please refer to chapter 20.
21.6 Weather observations
The file of weather observations provided here (Illinois_weather_1979-2011.txt) is by month and then by county for the state of Illinois from 1979 to 2011. Write the code to process this file to extract the data for Chicago (Cook County) into a single CSV or spreadsheet file. This process includes replacing the ‘Missing’ strings with empty strings and translating the percentage to a decimal. You may also consider what fields are repetitive (and therefore can be omitted or stored elsewhere). The proof that you’ve got it right occurs when you load the file into a spreadsheet, or you can also use Colaboratory’s file browser (the file icon at the very left) and double-click on it to see the data as a table.
There is some documentation at the end of the file, so you will need to stop processing the file when the first field of the line is “—”.
For reference, it’s always a good idea to look at a few lines (at least) of the raw file:
['"Notes"\t"Month"\t"Month Code"\t"County"\t"County Code"\tAvg Daily Max
Air Temperature (F)\tRecord Count for Daily Max Air Temp (F)\tMin Temp for
Daily Max Air Temp (F)\tMax Temp for Daily Max Air Temp (F)\tAvg Daily Min
Air Temperature (F)\tRecord Count for Daily Min Air Temp (F)\tMin Temp for
Daily Min Air Temp (F)\tMax Temp for Daily Min Air Temp (F)\tAvg Daily Max
Heat Index (F)\tRecord Count for Daily Max Heat Index (F)\tMin for Daily
Max Heat Index (F)\tMax for Daily Max Heat Index (F)\tDaily Max Heat Index
(F) % Coverage\n',
'\t"Jan"\t"1"\t"Adams County, IL"\t"17001"\t31.89\t19437\t
10.00\t68.90\t18.01\t19437\t-26.20\t50.30\tMissing\t0\tMissing\tMissing\
t0.00%\n',
'\t"Jan"\t"1"\t"Alexander County,
IL"\t"17003"\t41.07\t6138\t2.60\t73.20\t26.48\t6138\t
14.00\t60.30\tMissing\t0\tMissing\tMissing\t0.00%\n',
'\t"Jan"\t"1"\t"Bond County, IL"\t"17005"\t35.71\t6138\t-
2.70\t69.50\t22.18\t6138\t-17.90\t57.20\tMissing\t0\tMissing\tMissing\
t0.00%\n']21.6.1 Solving the problem with AI-generated code
This problem is mostly procedural, with little higher-level design, so it’s fairly suitable for an AI solution. As usual, we also need to be sure to include all of the necessary information clearly in the prompt.
21.6.2 Solutions and discussion
This is a much easier problem than those in some of the previous chapters, since we are really just doing some basic data cleaning. The main things to do are load the file, fix/discard some of the data fields, and then write the file as a CSV file.
The human solution
This solution is pretty simple: the input and output files are opened and the raw fields are read in, processed, and saved:
import csv
with (
open("Illinois_weather_1979-2011.txt") as infile,
open("chicago_weather_1979-2011.csv", "w") as outfile,
): # <-- Opens both input and output files
for row in csv.reader(infile, delimiter="\t"):
if row[0] == "---": # <-- Checks for beginning of documentation section
break
if "Cook" in row[3]:
del row[0:2] # <-- Removes first two fields of line
row = [
item.replace("Missing", "") for item in row
] # <-- Replaces “Missing” with an empty string
row[-1] = row[-1].replace(
"%", ""
) # <-- Removes % character from the last field
row[-1] = (
float(row[-1]) / 100 if str.isdigit(row[-1]) else row[-1]
) # <-- Convert percentage to a decimal value
csv.writer(outfile).writerow(row)As mentioned, the last few lines of this file are documentation, so we end processing when we hit the separator “—”. Since we are looking for Chicago (Cook County) data, we will only process a line if the “County” field contains “Cook”. Alternatively, we could look in the file and see that the county code for Cook County is “17031” and only process lines where that is in the “County Code” field. As we process the lines, the first thing we do is remove the first two fields—the first field (“Notes”) is empty, and second is the month name, which we don’t need since we have the month’s number in the following field.
The next two operations are changing “Missing” to an empty string in any field and then removing the % character and converting a text string of the percentage to a decimal number (e.g., “10” becomes 0,1).
The code finishes by writing the newly processed fields to the CVS file.
The AI solutions
Copilot was given the following prompt, which was based closely on the lab assignment:
The file of weather observations provided here (Illinois_weather_1979 2011.txt) is by month and then by county for the state of Illinois from 1979 to 2011. Write the code to process this file to extract the data for Chicago (Cook County) into a single CSV or spreadsheet file. This process includes replacing the ‘Missing’ strings with empty strings and translating the percentage to a decimal. You may also consider what fields are repetitive (and therefore can be omitted or stored elsewhere). Note: there is some documentation at the end of the file, so you will need to stop processing the file when the first field of the line is “—”.
Copilot offered the following plan, which makes sense:
- 1 Open the file Illinois_weather_1979-2011.txt for reading.
- 2 Create a new CSV file for writing the processed data.
- 3 Iterate through each line of the input file:
- a If the line starts with “—”, stop processing.
- b If the line contains data for Cook County (Chicago), process it:
- Replace “Missing” with an empty string.
- Convert percentage values to decimals.
- Write the processed data to the CSV file.
- 4 Close both files.
As expected, Copilot did fairly well with this task, offering a solution that almost does the job. It generated this code:
import csv
input_file = "Illinois_weather_1979-2011.txt"
output_file = "Chicago_weather_1979-2011.csv"
with open(input_file, "r") as infile, open(output_file, "w", newline="") as outfile:
reader = csv.reader(infile, delimiter="\t")
writer = csv.writer(outfile)
headers = next(reader)
writer.writerow(headers) # Write headers to the output file
for row in reader:
if row[0].startswith("---"):
break
if "Cook" in row[3]: # Assuming 'Cook' indicates Cook County (Chicago)
processed_row = [field.replace("Missing", "") for field in row]
processed_row = [
str(float(field.strip("%")) / 100) if "%" in field else field
for field in processed_row
]
writer.writerow(processed_row)This code is quite similar to the human version, with only a few differences. First, it handles headers nicely, copying them to the new file—something that the human version did not do. Second, it does the conversion of the percentages to decimals in one line, rather than two, and checks all fields rather than only the last one. Both of those differences are reasonable—even positive. Finally, all of the fields, even the empty “Notes” field, are preserved. This isn’t surprising, since removing/consolidating fields would be a value judgment.
The only problem is that when this code is run, the resulting file, Chicago_ weather_1979-2011.csv, only has the header. That’s because it checks for “Cook” in the entire row:
if 'Cook' in row:For that condition to be true, an entire field in the row would have to be exactly “Cook,” when in fact the rows we want contain a field “Cook County, IL”. The fix for this is to just specify that only row[3] contain “Cook”:
if 'Cook' in row[3]:With that one change, Copilot’s version works perfectly. Here again, we could spend a lot of time experimenting with the prompt to get a complete answer, but the efficient choice is to just make the three-character fix.
The prompt for Colaboratory was a bit different:
The file provided here (Illinois_weather_1979-2011.txt) is for the state of Illinois from 1979 to 2011. Write code to process this file to extract the data for Chicago (Cook County) into a single CSV. Replace the ‘Missing’ strings with empty strings and translate the percentage to a decimal. Use the input file headers to decide what fields are repetitive (and therefore can be omitted). Stop processing the file when the first field of the line is “—”.
The prompt was edited both since there is a 600-character limit in Colaboratory and because, with the original phrasing, the code generator assigned fields incorrectly. The current wording got around both problems:
# prompt: The file provided here (Illinois_weather_1979-2011.txt) is for the
# state of Illinois from 1979 to 2011. Write code to process this file
# to extract the data for Chicago (Cook County) into a single CSV.
# Replace the 'Missing' strings with empty strings and translate the
# percentage to a decimal. Use the input file headers to decide what
# fields are repetitive (and therefore can be omitted). Stop processing
# the file when the first field of the line is "---".
import csv
def process_illinois_weather_data():
"""Processes the Illinois weather data file and extracts Chicago data into a CSV."""
with open("Illinois_weather_1979-2011.txt", "r") as infile, open(
"chicago_weather.csv", "w", newline=""
) as outfile:
reader = csv.reader(infile, delimiter="\t")
writer = csv.writer(outfile)
header = next(reader)
writer.writerow(header)
for row in reader:
if row[0] == "---":
break
if "Cook" in row[3]:
new_row = []
for cell in row:
if cell == "Missing":
new_row.append("")
elif "%" in cell:
new_row.append(str(float(cell.replace("%", "")) / 100))
else:
new_row.append(cell)
writer.writerow(new_row)
process_illinois_weather_data()
print("Chicago weather data extracted to chicago_weather.csv")This approach is a bit different. While it preserves all of the fields just as the Copilot version did, it takes a different approach to processing a row. Rather than changing fields in a row, it instead processes each field in a row and then adds that field to a new row, which it writes to the file when the processing for the row is complete. This approach is arguably slightly less efficient since it creates a new list for each row, but in practice, the performance hit is probably slight.
As with the Copilot version, the code as suggested will write a file with only the header row—and for a very similar reason: in checking for Cook County results, it checks to see if row[1] == “Cook”. This is wrong both because the value would be “Cook County, IL”, as mentioned earlier, and because it checks the Month field, not the County field. The correction would be the same as before. The line should be
if 'Cook' in row[3]:Once that change is made, this code produces the correct result.
As mentioned, this task is fairly straightforward, and so it’s not as much of a challenge for the AI tools as some more complex tasks. The AI bots’ problems came in using the correct technique and field to check the county and in evaluating the fields for possible redundancy. Giving an explicit mapping and instructions in the prompt probably would have helped this, at the expense of making constructing the prompt more laborious.
Summary
- ETL is the process of getting data from one format, making sure that it’s consistent, and then putting it in a format you can use. ETL is the basic step in most data processing.
- Encoding can be problematic with text files, but Python lets you deal with some encoding problems when you load files.
- The most common way to handle a wide range of characters in simple text files is to encode them in the Unicode UTF-8 encoding.
- Delimited and CSV files are common, and the best way to handle them is with the csv module.
- Spreadsheet files (in Excel format) can be more complex than CSV files but can be read and written with third-party libraries.
- Currency symbols, punctuation, and null characters are among the most common data cleaning concerns; be on the watch for them.
- Cleaning and presorting your data file can make other steps faster.
- Writing data in the formats discussed is similar to reading them.
22 Data over the network
This chapter covers
- Fetching files via FTP/SFTP, SSH/SCP, and HTTPS
- Getting data via APIs
- Structured data file formats: JSON and XML
- Scraping data
You’ve seen how to deal with text-based data files. In this chapter, you use Python to move data files over the network. In some cases, those files might be text or spreadsheet files, as discussed in chapter 21, but in other cases, they might be in more structured formats and served from REST or SOAP APIs. Sometimes getting the data may mean scraping it from a website. This chapter discusses all of these situations and shows some common use cases.
22.1 Fetching files
Before you can do anything with data files, you have to get them. Sometimes this process is very easy, such as manually downloading a single zip archive, or maybe the files have been pushed to your machine from somewhere else. Quite often, however, the process is more involved. Maybe a large number of files need to be retrieved from a remote server, files need to be retrieved regularly, or the retrieval process is sufficiently complex to be a pain to do manually. In any of those cases, you might well want to automate fetching the data files with Python.
First, I want to be clear that using a Python script isn’t the only way, or always the best way, to retrieve files. The following sidebar offers more explanation of the factors I consider when deciding whether to use a Python script for file retrieval. Assuming that using Python does make sense for your particular use case, however, this section illustrates some common patterns you might employ.
Although using Python to retrieve files can work very well, it’s not always the best choice. In making a decision, you might want to consider two things:
- Are simpler options available? Depending on your operating system and your experience, you may find that simple shell scripts and command-line tools are simpler and easier to configure. If you don’t have those tools available or aren’t comfortable using them (or the people who will be maintaining them aren’t comfortable with them), you may want to consider a Python script.
- Is the retrieval process complex or tightly coupled with processing? Although those situations are never desirable, they can occur. My rule these days is that if a shell script requires more than a few lines, or if I have to think hard about how to do something in a shell script, it’s probably time to switch to Python.
22.1.1 Using Python to fetch files from an FTP server
File transfer protocol (FTP) has been around for a very long time, but it’s still a simple and easy way to share files when security isn’t a huge concern. To access an FTP server in Python, you can use the ftplib module from the standard library. The steps to follow are straightforward: create an FTP object, connect to a server, and then log in with a username and password (or, quite commonly, with a username of “anonymous” and an empty password).
To continue working with weather data, you can connect to the National Oceanic and Atmospheric Administration FTP server, as shown here:
import ftplib
ftp = ftplib.FTP('tgftp.nws.noaa.gov')
ftp.login()
#> '230 Login successful.'When you’re connected, you can use the ftp object to list and change directories:
ftp.cwd('data')
#> '250 Directory successfully changed.'ftp.nlst()
#> ['climate', 'fnmoc', 'forecasts', 'hurricane_products', 'ls_SS_services',
#> 'marine', 'nsd_bbsss.txt', 'nsd_cccc.txt', 'observations', 'products',
#> 'public_statement', 'raw', 'records', 'summaries', 'tampa',
#> 'watches_warnings', 'zonecatalog.curr', 'zonecatalog.curr.tar']Then you can fetch, for example, the latest METAR report for Chicago O’Hare International Airport:
x = ftp.retrbinary('RETR observations/metar/decoded/KORD.TXT',
open('KORD.TXT', 'wb').write)
#> '226 Transfer complete.'The ftp.retrbinary method takes two parameters—the path to the file on the remote server and a method to handle that file’s data when you receive it on your end; in this case, that method is the write method of a file opened for binary writing with the same name. When you look at KORD.TXT, you see that it contains the downloaded data:
x = ftp.retrbinary('RETR observations/metar/decoded/KORD.TXT', open('KORD.
TXT', 'wb').write)
open('KORD.TXT', 'r').readlines()
#> ["CHICAGO O'HARE INTERNATIONAL, IL, United States (KORD)
#> ➥41-59N 087-55W 200M\n",
#> 'Aug 19, 2024 - 10:51 PM EDT / 2024.08.20 0251 UTC\n',
#> 'Wind: from the NNE (030 degrees) at 13 MPH (11 KT):0\n',
#> 'Visibility: 10 mile(s):0\n',
#> 'Sky conditions: partly cloudy\n',
#> 'Temperature: 68.0 F (20.0 C)\n',
#> 'Dew Point: 55.9 F (13.3 C)\n',
#> 'Relative Humidity: 65%\n',
#> 'Pressure (altimeter): 30.17 in. Hg (1021 hPa)\n',
#> 'Pressure tendency: 0.03 inches (1.0 hPa) higher than three hours ago\n',
#> 'ob: KORD 200251Z 03011KT 10SM SCT039 SCT050 20/13 A3017
#> ➥RMK AO2 SLP212 T02000133 53010\n',
#> 'cycle: 3\n']You can also use ftplib to connect to servers using Transport Layer Security (TLS) encryption by using FTP_TLS instead of FTP:
ftp = ftplib.FTPTLS('tgftp.nws.noaa.gov')22.1.2 Fetching files with SFTP
If the data requires more security, such as in a corporate context in which business data is being transferred over the network, it’s fairly common to use SFTP. SFTP is a fullfeatured protocol that allows file access, transfer, and management over a Secure Shell (SSH) connection. Even though SFTP stands for SSH file transfer protocol and FTP stands for file transfer protocol, the two aren’t related. SFTP isn’t a reimplementation of FTP on SSH but a fresh design specifically for SSH.
Using SSH-based transfers is attractive both because SSH is already the de facto standard for accessing remote servers and because enabling support for SFTP on a server is fairly easy (and quite often on by default).
Python doesn’t have an SFTP/SCP client module in its standard library, but a community-developed library called paramiko manages SFTP operations as well as SSH connections. To use paramiko, the easiest thing is to install it via pip. If the National Oceanic and Atmospheric Administration site mentioned earlier in this chapter were using SFTP (which it doesn’t, so this code won’t work!), the SFTP equivalent of the previous code would be
import paramiko
t = paramiko.Transport((hostname, port))
t.connect(username, password)
sftp = paramiko.SFTPClient.from_transport(t)It’s also worth noting that, although paramiko supports running commands on a remote server and receiving its outputs, just like a direct ssh session, it doesn’t include an scp function. This function is rarely something you’ll miss; if all you want to do is move a file or two over an ssh connection, a command-line scp utility usually makes the job easier.
22.1.3 Retrieving files over HTTP/HTTPS
The last common option for retrieving data files that I discuss in this chapter is getting files over an HTTP or HTTPS connection. This option is probably the easiest of all the options; you are in effect retrieving your data from a web server, and support for accessing web servers is very widespread. Again, in this case, you may not need to use Python. Various command-line tools retrieve files via HTTP/HTTPS connections and have most of the capabilities you might need. The two most common of these tools are wget and curl. If you have a reason to do the retrieval in your Python code, however, that process isn’t much harder. The requests library is by far the easiest and most reliable way to access HTTP/HTTPS servers from Python code. Again, requests is easiest to install with pip install requests. When you have requests installed, fetching a file is straightforward: import requests and use the correct HTTP verb (usually, GET) to connect to the server and return your data.
The following example code fetches the monthly temperature data for Heathrow Airport since 1948—a text file that’s served via a web server. If you want to, you can put the URL in your browser, load the page, and then save it. If the page is large or you have a lot of pages to get, however, it’s easier to use code like the following:
import requests
response = requests.get("http://www.metoffice.gov.uk/pub/data/weather/uk/climate/stationdata/heathrowdata.txt")The response will have a fair amount of information, including the header returned by the web server, which can be helpful in debugging if things aren’t working. The part of the response object you’ll most often be interested in, however, is data returned. To retrieve this data, you want to access the response’s text property, which contains the response body as a string, or the content property, which contains the response body as bytes:
print(response.text)
# Heathrow (London Airport)
# Location 507800E 176700N, Lat 51.479 Lon -0.449, 25m amsl
# Estimated data is marked with a * after the value.
# Missing data (more than 2 days missing in month) is marked by ---.
# Sunshine data taken from an automatic Kipp & Zonen sensor marked with a #, otherwise sunshine data taken from a Campbell Stokes recorder.
# yyyy mm tmax tmin af rain sun
# degC degC days mm hours
# 1948 1 8.9 3.3 --- 85.0 ---
# 1948 2 7.9 2.2 --- 26.0 ---
# 1948 3 14.2 3.8 --- 14.0 ---
# 1948 4 15.4 5.1 --- 35.0 ---
# 1948 5 18.1 6.9 --- 57.0 ---
# 1948 6 19.1 10.3 --- 67.0 ---
# 1948 7 21.7 12.0 --- 21.0 ---
# ...Typically, you’d write the response text to a file for later processing, but depending on your needs, you might first do some cleaning or even process it immediately.
If you’re working with the example data file and want to break each line into separate fields, how might you do that? What other processing would you expect to do? Try writing some code to retrieve this file and calculate the average annual rainfall or (for more of a challenge) the average maximum and minimum temperature for each year.
22.2 Fetching data via an API
Serving data by way of an API is quite common, following a trend toward decoupling applications into services that communicate via APIs. APIs can work in several ways, but they commonly operate over regular HTTP/HTTPS protocols using the standard HTTP verbs: GET, POST, PUT, and DELETE. Fetching data this way is very similar to retrieving a file, as in section 22.1.3, but the data isn’t in a static file. Instead of the application serving static files that contain the data, it queries some other data source and then assembles and serves the data dynamically on request.
Although there’s a lot of variation in the ways that an API can be set up, one of the most common is a REpresentational State Transfer (RESTful) interface that operates over the same HTTP/HTTPS protocols as the web. There are endless variations on how an API might work, but commonly, data is fetched by using a GET request, which is what your web browser uses to request a web page. When you’re fetching via a GET request, the parameters to select the data you want are often appended to the URL in a query string.
We can test this by getting the current weather in Chicago from Open-Meteo.com, using https://mng.bz/BX9g as your URL. The items after the ? are query string parameters that specify the latitude and longitude of the location and the two fields that we are requesting: temperature and wind speed. For this API, the information will be returned in JSON, which I discuss in section 22.3.1. Notice that the elements of the query string are separated by ampersands (&).
When you know the URL to use, you can use the requests library to fetch data from an API and either process it on the fly or save it to a file for later processing. The simplest way to do this is exactly like retrieving a file:
response = requests.get("https://api.open-meteo.com/v1/forecast?latitude=41.882&longitude=-87.623¤t=temperature_2m,wind_speed_10m")
weather = response.text
weather
#> {"latitude":41.879482,"longitude":-87.64975,"generationtime_ms":0.026941299
#> 438476562,"utc_offset_seconds":0,"timezone":"GMT","timezone_abbreviation":
#> "GMT","elevation":185.0,"current_units":{"time":"iso8601","interval":
#> "seconds","temperature_2m":"°C","wind_speed_10m":"km/h"},"current":{"time":
#> "2024-09-02T02:00","interval":900,"temperature_2m":16.9,
#> "wind_speed_10m":13.3}}Keep in mind that, ideally, you should escape spaces and most punctuation in your query parameters, because those elements aren’t allowed in URLs. It’s not absolutely necessary, though, because the requests library and many browsers automatically do the escaping on URLs.
For a final example, suppose that you want to grab the crime data for Chicago between noon and 1 P.M. on January 10, 2017. The way that the API works, you specify a date range with the query string parameters of $where=date between <start datetime> and <end datetime>, where the start and end datetimes are quoted in ISO format. So the URL for getting that 1 hour of Chicago crime data would be https://mng.bz/N1ad:
result = requests.get("https://data.cityofchicago.org/resource/6zsd-86xi.json?$where=date between '2024-01-10T12:01:00' and
'2024-01-10T12:10:00'")
result.json()
# [{'id': '13334404',
# 'case_number': 'JH111115',
# 'date': '2024-01-10T12:01:00.000',
# 'block': '009XX W WINONA ST',
# 'iucr': '2020',
# 'primary_type': 'NARCOTICS',
# 'description': 'POSSESS - AMPHETAMINES',
# 'location_description': 'APARTMENT',
# 'arrest': True,
# 'domestic': False,
# 'beat': '2024',
# 'district': '020',
# 'ward': '48',
# 'community_area': '3',
# 'fbi_code': '18',
# 'x_coordinate': '1169132',
# 'y_coordinate': '1934325',
# 'year': '2024',
# 'updated_on': '2024-01-18T15:41:44.000',
# 'latitude': '41.975305734',
# 'longitude': '-87.653416364',
# 'location': {'type': 'Point', 'coordinates':
# [-87.653416364, 41.975305734]},
# 'location_address': '',
# 'location_city': '',
# 'location_state': '',
# 'location_zip': ''},
# {'id': '13334420',
# 'case_number': 'JH111020',
# 'date': '2024-01-10T12:01:00.000',
# 'block': '009XX W WINONA ST',
# 'iucr': '1330',
# 'primary_type': 'CRIMINAL TRESPASS',
# 'description': 'TO LAND',
# 'location_description': 'APARTMENT',
# 'arrest': True,
# 'domestic': False,
# 'beat': '2024',
# 'district': '020',
# 'ward': '48',
# 'community_area': '3',
# 'fbi_code': '26',
# 'x_coordinate': '1169132',
# 'y_coordinate': '1934325',
# 'year': '2024',
# 'updated_on': '2024-01-18T15:41:44.000',
# 'latitude': '41.975305734',
# 'longitude': '-87.653416364',
# 'location': {'type': 'Point', 'coordinates':
# [-87.653416364, 41.975305734]},
# 'location_address': '',
# 'location_city': '',
# 'location_state': '',
# 'location_zip': ''},
# ... # <-- Several records omitted
# ]In the example, some characters aren’t welcome in URLs, such as the space characters. This is another situation in which the requests library makes good on its aim of making things easier for the user, because, before it sends the URL, it takes care of quoting it properly. The URL that the request actually sends is https://mng.bz/dXvX.
Note that all of the spaces have been quoted with %20 without your even needing to think about it.
Write some code to fetch some data from the city of Chicago website. Look at the fields mentioned in the results and see whether you can select records based on another field in combination with the date range.
22.3 Structured data formats
Although APIs sometimes serve plain text, it’s much more common for data served from APIs to be served in a structured file format. The two most common file formats are JSON and XML. Both of these formats are built on plain text but structure their contents so that they’re more flexible and able to store more complex information.
22.3.1 JSON data
JSON, which stands for JavaScript Object Notation, dates to 1999. It consists of only two structures: key-value pairs, called structures, that are very similar to Python dictionaries and ordered lists of values, called arrays, that are very much like Python lists.
Keys can be only strings in double quotes, and values can be strings in double quotes, numbers, true, false, null, arrays, or other structures. These elements make JSON a lightweight way to represent most data in a way that’s easily transmitted over the network and also fairly easy for humans to read. JSON is so common that most languages have features to translate JSON to and from native data types. In the case of Python, that feature is the json module, which became part of the standard library with version 2.6. The original externally maintained version of the module is available as simplejson, which is still available. In Python 3, however, it’s far more common to use the standard library version, unless you need a feature only supported in simplejson, like the decimal number type.
The data you retrieved from the weather and the city of Chicago APIs in section 22.2 is in JSON format. To send JSON across the network, the JSON object needs to be serialized—that is, transformed into a sequence of bytes. So, although the batch of data you retrieved from the weather and Chicago crime APIs looks like JSON, in fact, it’s just a byte string representation of a JSON object. To transform that byte string into a real JSON object and translate it into a Python dictionary, you need to use the JSON loads() function. If you want to get the weather report, for example, you can do that just as you did previously, but this time you’ll convert it to a Python dictionary:
import json
import requests
response = requests.get("https://api.open-meteo.com/v1/forecast?latitude=41.882&longitude=-87.623¤t=temperature_2m,wind_speed_10m")
weather = json.loads(response.text)
weather
# {'latitude': 41.879482,
# 'longitude': -87.64975,
# 'generationtime_ms': 0.025987625122070312,
# 'utc_offset_seconds': 0,
# 'timezone': 'GMT',
# 'timezone_abbreviation': 'GMT',
# 'elevation': 185.0,
# 'current_units': {'time': 'iso8601',
# 'interval': 'seconds',
# 'temperature_2m': '°C',
# 'wind_speed_10m': 'km/h'},
# 'current': {'time': '2024-09-02T02:15',
# 'interval': 900,
# 'temperature_2m': 16.9,
# 'wind_speed_10m': 13.8}}weather['current']['temperature_2m']
#> 16.9Note that the call to json.loads() is what takes the string representation of the JSON object and transforms, or loads, it into a Python dictionary. Also, a json.load() function will read from any file-like object that supports a read method.
If you print a dictionary’s representation, it can be very hard to make sense of what’s going on. Improved formatting, also called pretty printing, can make data structures much easier to understand. Use the Python prettyprint module to see what’s in the example dictionary:
from pprint import pprint as pp
pp(weather)
# {'current': {'interval': 900,
# 'temperature_2m': 16.9,
# 'time': '2024-09-02T02:15',,
# 'wind_speed_10m': 13.8},
# 'current_units': {'interval': 'seconds',
# 'temperature_2m': '°C',
# 'time': 'iso8601',
# 'wind_speed_10m': 'km/h'},
# 'elevation': 185.0,
# 'generationtime_ms': 0.025987625122070312,
# 'latitude': 41.879482,
# 'longitude': -87.64975,
# 'timezone': 'GMT',
# 'timezone_abbreviation': 'GMT',
# 'utc_offset_seconds': 0}Both load functions can be configured to control how to parse and decode the original JSON to Python objects, but the default translation is listed in table 22.1.
| JSON | Python |
|---|---|
object |
dict |
array |
list |
string |
str |
number (int) |
int |
number (real) |
float |
true |
True |
false |
False |
null |
None |
In this section, you used the requests library to retrieve the JSON-formatted data and then used the json.loads() method to parse it into a Python object. This technique works fine, but because the requests library is used so often for exactly this purpose, the library provides a shortcut: the response object actually has a json() method that does that conversion for you. So, in the example, instead of
weather = json.loads(response.text)you could have used
weather = response.json()The result is the same, but the code is simpler, more readable, and more Pythonic.
If you want to write JSON to a file or serialize it to a string, the reverse of load() and loads() is dump() and dumps(). json.dump() takes a file object with a write() method as a parameter, and json.dumps() returns a string. In both cases, the encoding to a JSON-formatted string can be highly customized, but the default is still based on table 22.1. So if you want to write your weather report to a JSON file, you could do the following:
outfile = open("weather_01.json", "w")
json.dump(weather, outfile) # <-- Writes the string to outfile
outfile.close()json.dumps(weather) # <-- Returns the string'{"latitude": 41.879482, "longitude": -87.64975, "generationtime_ms": '
'23.406624794006348, "utc_offset_seconds": 0, "timezone": "GMT", '
'"timezone_abbreviation": "GMT", "elevation": 185.0, "current_units": '
'{"time": "iso8601", "interval": "seconds", "temperature_2m": "\\u00b0C", '
'"wind_speed_10m": "km/h"}, "current": {"time": "2025-11-25T15:00", '
'"interval": 900, "temperature_2m": 8.5, "wind_speed_10m": 4.9}}'As you can see, the entire object has been encoded as a single string. With json.dump (with no “s” on the end) that string is written to the specified file. With the “s,” json .dumps returns the serialized string. Here again, it might be handy to format the string in a more readable way, just as you did by using the pprint module. To do so easily, use the indent parameter with the dump or dumps function:
print(json.dumps(weather, indent=2)){
"latitude": 41.879482,
"longitude": -87.64975,
"generationtime_ms": 23.406624794006348,
"utc_offset_seconds": 0,
"timezone": "GMT",
"timezone_abbreviation": "GMT",
"elevation": 185.0,
"current_units": {
"time": "iso8601",
"interval": "seconds",
"temperature_2m": "\u00b0C",
"wind_speed_10m": "km/h"
},
"current": {
"time": "2025-11-25T15:00",
"interval": 900,
"temperature_2m": 8.5,
"wind_speed_10m": 4.9
}
}You should be aware, however, that if you use repeated calls to json.dump()to write a series of objects to a file, the result is a series of legal JSON-formatted strings, but the contents of the file as a whole is not a legal JSON-formatted object, and attempting to read and parse the entire file by using a single call to json.load() will fail. If you have more than one object that you’d like to encode as a single JSON object, you need to put all those objects into a list (or, better yet, an object or structure) and then encode that item to the file.
If you have two or more items of weather data that you want to store in a file as JSON, you have to make a choice. You could use json.dump() once for each object, which would result in a file containing JSON-formatted strings. If you assume that weather_ list is a list of weather-report objects, the code might look like the following:
outfile = open("weather_list.json", "w")
for report in weather_list:
outfile.write(json.dumps(report) + "\n")
outfile.close()If you do this, then you need to load each line as a separate JSON-formatted object:
weather_list2 = []
for line in open("weather_list.json"):
weather_list2.append(json.loads(line))As an alternative, you could put the list into a single JSON object. Because there’s a possible vulnerability with top-level arrays in JSON, the recommended way is to put the array in a dictionary:
outfile = open("weather_obj.json", "w")
weather_obj = {"reports": weather_list, "count": 2}
json.dump(weather_obj, outfile)
outfile.close()With this approach, you can use one operation to load the JSON-formatted object from the file:
with open("weather_obj.json") as infile:
weather_obj = json.load(infile))The second approach is fine if the size of your JSON files is manageable, but it may be less than ideal for very large files, because handling errors may be a bit harder and you may run out of memory.
Modify the code you wrote in section 22.2 to fetch the Chicago crime data. Then convert the fetched data from a JSON-formatted string to a Python object. Next, see whether you can save the crime events as a series of separate JSON strings in one file and as one JSON object in another file. Then see what code is needed to load each file.
22.3.2 XML data
XML has been around since the end of the 20th century. XML uses an angle-bracket tag notation similar to HTML, and elements are nested within other elements to form a tree structure. XML was intended to be readable by both machines and humans, but XML is often so verbose and complex that it’s very difficult for people to understand. Nevertheless, because XML is an established standard, it’s quite common to find data in XML format. And although XML is machine readable, it’s very likely that you’ll want to translate it into something a bit easier to deal with.
To take a look at some XML data, let’s fetch the XML version of the World Bank’s population data for Chile:
result = requests.get(
"https://api.worldbank.org/v2/country/CL/indicator/SP.POP.TOTL?format=xml")
pop_data = result.text[3:] # <-- Skips extra characters
print(pop_data)
# <?xml version="1.0" encoding="utf-8"?>
# <wb:data page="1" pages="2" per_page="50" total="64" sourceid="2"
# lastupdated="2024-06-28" xmlns:wb="http://www.worldbank.org">
# <wb:data>
# <wb:indicator id="SP.POP.TOTL">Population, total</wb:indicator>
# <wb:country id="CL">Chile</wb:country>
# <wb:countryiso3code>CHL</wb:countryiso3code>
# <wb:date>2023</wb:date>
# <wb:value>19629590</wb:value>
# <wb:unit />
# <wb:obs_status />
# <wb:decimal>0</wb:decimal>
# </wb:data>
# <wb:data>
# <wb:indicator id="SP.POP.TOTL">Population, total</wb:indicator>
# <wb:country id="CL">Chile</wb:country>
# <wb:countryiso3code>CHL</wb:countryiso3code>
# <wb:date>2022</wb:date>
# <wb:value>19603733</wb:value>
# <wb:unit />
# <wb:obs_status />
# <wb:decimal>0</wb:decimal>
# </wb:data>
# ... # <-- Several records omitted
# </wb:data> This example is just the first section of the document, with most of the data omitted. Even so, it illustrates some of the concerns you typically find in XML data. In particu- lar, you can see the verbose nature of the protocol, with the tags in some cases taking more space than the value contained in them. This sample also shows the nested or tree structure common in XML, as well as the common use of a sizeable header of metadata before the actual data begins. On a spectrum from simple to complex for data files, you could think of CSV or delimited files as being at the simple end and XML at the complex end.
Finally, this file illustrates another feature of XML that makes pulling data a bit more of a challenge. XML supports the use of attributes to store data as well as the text values within the tags. So, if you look at the first wb:data element at the top of this sample, you see that this element contains its metadata as tags and then also contains the other data elements. To know which way any given bit of data will be handled, you need to carefully inspect the data or study a specification document.
This kind of complexity can make simple data extraction from XML more of a challenge. You have several ways to handle XML. The Python standard library comes with modules that parse and handle XML data, but none of them are particularly convenient for simple data extraction.
For simple data extraction, the handiest utility I’ve found is a library called xmltodict, which parses your XML data and returns a dictionary that reflects the tree. In fact, behind the scenes, it uses the standard library’s Expat XML parser, parses your XML document into a tree, and uses that tree to create the dictionary. As a result, xmltodict can handle whatever the parser can, and it’s also able to take a dictionary and “unparse” it to XML if necessary, making it a very handy tool. Over several years of use, I found this solution to be up to all my XML handling needs. To get xmltodict, you can again use pip install xmltodict.
To convert the XML to a dictionary, you can import xmltodict and use the parse method on an XML-formatted string:
import xmltodict
population = xmltodict.parse(pop_data)
populationIn this case, for compactness, pass the contents of the file directly to the parse method. After being parsed, this data object is an ordered dictionary with the same values it would have if it had been loaded from this JSON:
{'wb:data': {'@page': '1',
'@pages': '2',
'@per_page': '50',
'@total': '64',
'@sourceid': '2',
'@lastupdated': '2024-06-28',
'@xmlns:wb': 'http://www.worldbank.org',
'wb:data': [{'wb:indicator': {'@id': 'SP.POP.TOTL',
'#text': 'Population, total'},
'wb:country': {'@id': 'CL', '#text': 'Chile'},
'wb:countryiso3code': 'CHL',
'wb:date': '2023',
'wb:value': '19629590',
'wb:unit': None,
'wb:obs_status': None,
'wb:decimal': '0'},
{'wb:indicator': {'@id': 'SP.POP.TOTL', '#text': 'Population, total'},
'wb:country': {'@id': 'CL', '#text': 'Chile'},
'wb:countryiso3code': 'CHL',
'wb:date': '2022',
'wb:value': '19603733',
'wb:unit': None,
'wb:obs_status': None,
'wb:decimal': '0'},
… # <-- Several records omitted
]
}
}Notice that the attributes have been pulled out of the tags but with an @ prepended to indicate that they were originally attributes of their parent tag. If an XML node has both a text value and a nested element in it, notice that the key for the text value is “#text”, as in the “wb-indicator” element in each “wb-data” element. If an element is repeated, it becomes a list, as the list of wb-data elements in this sample.
Because dictionaries and lists—even nested dictionaries and lists—are fairly easy to deal with in Python, using xmltodict is an effective way to handle most XML. In fact, I’ve used it for the past several years in production on a variety of XML documents and never had a problem.
Write the code to pull the XML data for Chile’s population from https://mng.bz/Ea6R. Then use xmltodict to parse the XML into a Python dictionary and report how much Chile’s population has changed in the past 25 years.
22.4 Scraping web data
In some cases, the data is on a website but for whatever reason isn’t available anywhere else. In those situations, it may make sense to collect the data from the web pages themselves through a process called crawling or scraping.
Before saying anything more about scraping, let me make a disclaimer: scraping or crawling websites that you don’t own or control is at best a legal grey area, with a host of inconclusive and contradictory considerations involving things such as the terms of use of the site, the way in which the site is accessed, and the use to which the scraped data is put. Unless you have control of the site you want to scrape, the answer to the question “Is it legal for me to scrape this site?” usually is “It depends.”
If you do decide to scrape a production website, you also need to be sensitive to the load you’re putting on the site. Although an established, high-traffic site might well be able to handle anything you can throw at it, a smaller, less active site might be brought to a standstill by a series of continuous requests. At the very least, you need to be careful that your scraping doesn’t turn into an inadvertent denial-of-service attack.
Conversely, I’ve worked in situations in which it was actually easier to scrape our own website to get some needed data than it was to go through corporate channels. Although scraping web data has its place, it’s too complex for full treatment here. In this section, I present a very simple example to give you a general idea of the basic method and follow up with suggestions to pursue in more complex cases.
Scraping a website consists of two parts: fetching the web page and extracting the data from it. Fetching the page can be done via requests and is fairly simple. Consider the code of a very simple web page with only a little content and no CSS or JavaScript, as the one shown in the following listing.
Listing 22.1 File test.html
<!DOCTYPE HTML PUBLIC "-//IETF//DTD HTML//EN">
<html> <head>
<title>Title</title>
</head>
<body>
<h1>Heading 1</h1>
This is plan text, and is boring
<span class="special">this is special</span>
Here is a <a href="http://bitbucket.dev.null">link</a>
<hr>
<address>Ann Address, Somewhere, AState 00000
</address>
</body> </html>Suppose that you’re interested in only a couple of kinds of data from this page: anything in an element with a class name of “special” and any links. You can process the file by searching for the strings ‘class=“special”’ and “<a href>” and then write code to pick out the data from there, but even using regular expressions, this process will be tedious, bug prone, and hard to maintain. It’s much easier to use a library that knows how to parse HTML, such as Beautiful Soup. If you want to try the following code and experiment with parsing HTML pages, you can use pip install bs4.
When you have Beautiful Soup installed, parsing a page of HTML is simple. For this example, assume that you’ve already retrieved the web page (presumably, using the requests library), so you’ll just parse the HTML.
The first step is to load the text and create a Beautiful Soup parser:
import bs4
html = open("test.html").read()
bs = bs4.BeautifulSoup(html, "html.parser")This code is all it takes to parse the HTML into the parser object bs. A Beautiful Soup parser object has a lot of cool tricks, and if you’re working with HTML at all, it’s really worth your time to experiment a bit and get a feel for what it can do for you. For this example, you look at only two things: extracting content by HTML tag and getting data by class.
First, find the link. The HTML tag for a link is <a> (Beautiful Soup by default converts all tags to lowercase), so to find all link tags, you can use the “a” as a parameter and call the bs object itself:
a_list = bs("a")
print(a_list)
[<a href="http://bitbucket.dev.null">link</a>]Now you have a list of all (one in this case) of the HTML link tags. If that list is all you get, that’s not so bad, but in fact, the elements returned in the list are also parser objects and can do the rest of the work of getting the link and text for you:
a_item = a_list[0]
a_item.text
#> 'link'
a_item["href"]
#> 'http://bitbucket.dev.null'The other feature you’re looking for is anything with a class of “special”, which you can extract by using the parser’s select method as follows:
special_list = bs.select(".special")
print(special_list)
#> [<span class="special">this is special</span>]
special_item = special_list[0]
special_item.text
#> 'this is special'
special_item["class"]
#> ['special']Because the items returned by the tag or by the select method are themselves parser objects, you can nest them, which allows you to extract just about anything from HTML or even XML.
Given the file forecast.html (which you can create in the notebook for this chapter, also shown bin listing 22.2), write a script using Beautiful Soup that extracts the data and saves it as a CSV file.
Listing 22.2 File forecast.html
Listing 22.2 File forecast.html
<html>
<body>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Tonight</b></div>
<div class="grid col-75 forecast-text">A slight chance of showers and thunderstorms before 10pm. Mostly cloudy, with a low around 66. West southwest wind around 9 mph. Chance of precipitation is 20%. New rainfall amounts between a tenth and quarter of an inch possible.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Friday</b></div>
<div class="grid col-75 forecast-text">Partly sunny. High near 77, with temperatures falling to around 75 in the afternoon. Northwest wind 7 to 12 mph, with gusts as high as 18 mph.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Friday Night</b></div>
<div class="grid col-75 forecast-text">Mostly cloudy, with a low around 63. North wind 7 to 10 mph.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Saturday</b></div>
<div class="grid col-75 forecast-text">Mostly sunny, with a high near 73. North wind around 10 mph.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Saturday Night</b></div>
<div class="grid col-75 forecast-text">Partly cloudy, with a low around 63. North wind 5 to 10 mph.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Sunday</b></div>
<div class="grid col-75 forecast-text">Mostly sunny, with a high near 73.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Sunday Night</b></div>
<div class="grid col-75 forecast-text">Mostly cloudy, with a low around 64.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Monday</b></div>
<div class="grid col-75 forecast-text">Mostly sunny, with a high near 74.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Monday Night</b></div>
<div class="grid col-75 forecast-text">Mostly clear, with a low around 65.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Tuesday</b></div>
<div class="grid col-75 forecast-text">Sunny, with a high near 75.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Tuesday Night</b></div>
<div class="grid col-75 forecast-text">Mostly clear, with a low around 65.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Wednesday</b></div>
<div class="grid col-75 forecast-text">Sunny, with a high near 77.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Wednesday Night</b></div>
<div class="grid col-75 forecast-text">Mostly clear, with a low around 67.</div>
</div>
<div class="row row-forecast">
<div class="grid col-25 forecast-label"><b>Thursday</b></div>
<div class="grid col-75 forecast-text">A chance of rain showers after 1pm. Mostly sunny, with a high near 81. Chance of precipitation is 30%.</div>
</div>
</body>
</html>22.5 Tracking the weather
Use the API described in section 22.2 to gather a history of the mean temperatures of a location (you can use Chicago or supply the latitude and longitude for a different location) for a month. You will need to use a slightly different URL for the archive API: “https://archive-api.open-meteo.com/v1/era5?latitude=<latitude>&longitude =<longitude>&start_date=<YYYY-MM-DD>&end_date=<YYYY-MM-DD>&daily= temperature_2m_mean” like this:
"https://archive-api.open-meteo.com/v1/era5?latitude=41.879&longitude=-87.64975&start_date=2024-07-01&end_date=2024-07-31&daily=temperature_2m_mean"Notice that you will get two lists: a list of the dates and a matching list of the mean temperatures in Celsius for those dates. Transform the data so that you can load it into a spreadsheet and graph it.
22.5.1 Solving the problem with AI-generated code
There is no complex design component in this lab. It requires a good knowledge of accessing an HTTP API and handling the JSON returned. As such, it’s something that AI should be good at.
22.5.2 Solutions and discussion
This problem is a matter of retrieving a file from an HTTP server and then processing the JSON returned to join two parallel lists.
The human solution
This solution takes a simple way through the problem, using the requests library to fetch and process the data and save the processed data to a file:
# **HINT:** Remember that spreadsheets can read CSV files, so you don't need to create an Excel file.
## Human solution
import requests
import csv
response = requests.get("https://archive-api.open-meteo.com/v1/era5?latitude=41.879&longitude=-87.64975&start_date=2024-07-01&end_date=2024-07-31&daily=temperature_2m_mean")
weather_history = response.json() # <-- The response object 's json() method returns data as JSON.
weather_history
weather_data = zip(weather_history["daily"]["time"], weather_history["daily"]["temperature_2m_mean"])
list(weather_data) # <-- The zip() function returns an iterator of the combined lists.
with open("weather_history.csv", "w", newline="") as file:
writer = csv.writer(file)
writer.writerow(["Date", "Temperature"])
writer.writerows(weather_data) # <-- The writerows() method writes all rows the from iterator weather_data.This solution takes advantage of the fact that requests can return the data as JSON, avoiding the need to parse the text into JSON. The two lists are zipped together, and the resulting data is written as a CSV file. One quirk in using zip() is that it returns an iterator, not a list. This isn’t a problem for the writerows() method, but if you want to look at the results, you need to use the list() function to turn it into a list.
The AI solutions
The prompt used for Copliot was really simplified from the lab problem, and the field names were specified:
The URL https://archive-api.open-meteo.com/v1/era5?latitude=41.879&longitude
=-87.64975&start_date=2024-07-
➥01&end_date=2024-07-31&daily=temperature_2m_mean will return one day's
weather data for Chicago in JSON format. The data will contain a "daily"
field with two lists, "time" and "temperature_2m_mean". Download the data
and save the time and temperature_2m_mean fields into a CSV file that can
later be loaded into a spreadsheet and graphed.This prompt was created to avoid confusion on field names and to remove any nonessential information. The resulting code was quite useful:
import requests
import csv
# URL to fetch the weather data
url = "https://archive-api.open-meteo.com/v1/era5?latitude=41.879&longitude=
-87.64975&start_date=2024-07-01
➥&end_date=2024-07-31&daily=temperature_2m_mean"
# Fetch the data
response = requests.get(url)
data = response.json()
# Extract the relevant fields
time_data = data['daily']['time']
temperature_data = data['daily']['temperature_2m_mean']# Output CSV file
output_file = 'Chicago_weather_July_2024.csv'
# Write the data to the CSV file
with open(output_file, 'w', newline='') as csvfile:
writer = csv.writer(csvfile)
writer.writerow(['Date', 'Temperature (°C)']) # Write the header
for time, temperature in zip(time_data, temperature_data):
writer.writerow([time, temperature])This code is fine and works as specified with no changes. It even iterates over the data to write the CSV file, while the human version processes the whole batch of data into a list that is written in one operation. Since the number of elements is known in this case, the memory-saving precaution isn’t really needed, but it’s interesting to see it. One that is a bit superfluous is the use of the zip() function to combine the two lists. While this is needed in my version to create a list of the data pairs, in this case, the pairs are manually joined into a list as each row is written, so they could have been read from the two lists directly. I would say that zipping them does make the code a bit more natural in Python. Otherwise, it’s very much like my version and works well as is.
Colaboratory was given the same prompt:
The URL https://archive-api.open-meteo.com/v1/era5?latitude=41.879
➥&longitude=-87.64975&start_date=2024-07-01
➥&end_date=2024-07-31&daily=temperature_2m_mean will return one day's
weather data for Chicago in JSON format. The data will contain a "daily"
field with two lists, "time" and "temperature_2m_mean". Download the data
and save the time and temperature_2m_mean fields into a CSV file that can
later be loaded into a spreadsheet and graphed.This prompt worked well for Colaboratory as well, although it took a somewhat different approach:
import csv
import requests
def download_and_save_chicago_weather():
"""Downloads Chicago weather data and saves it to a CSV file."""
url = "https://archive-api.open-meteo.com/v1/era5?latitude=41.879&longitude
=-87.64975
➥&start_date=2024-07-01&end_date=2024-07-31&daily=temperature_2m_mean"
response = requests.get(url)
if response.status_code == 200:
weather_data = response.json()
if "daily" in weather_data:
daily_data = weather_data["daily"]
if "time" in daily_data and "temperature_2m_mean" in daily_data:
with open('chicago_weather_july.csv', 'w', newline='') as outfile:
writer = csv.writer(outfile)
writer.writerow(['Date', 'Mean Temperature (°C)'])➥ # Write header row
for i in range(len(daily_data["time"])):
writer.writerow([daily_data["time"][i], daily_
data["temperature_2m_mean"][i]])
print("Chicago weather data for July 2024 saved to
➥chicago_weather_july.csv")
else:
print("Error: 'time' or 'temperature_2m_mean' not found
➥in daily data.")
else:
print("Error: 'daily' field not found in weather data.")
else:
print("Error downloading data:", response.status_code)download_and_save_chicago_weather()First, this code has the actual functionality in a function, download_and_save_chicago_ weather(), which is a nice touch. There isn’t much difference in the way the data is retrieved, but there is a fair bit of error checking. This is nice to have, particularly when it’s generated with no additional programmer effort, but it’s not particularly Pythonic. Instead of using if statements and the print function, it would have been more idiomatic to use exceptions, most commonly a KeyError for the cases where a field didn’t exist in the data and possibly some form of OSError for a failure to download the data (although probably an error will be raised by the requests library before the code gets that far).
Second, this code doesn’t zip() the two lists together but instead uses a for loop based on the length of the lists to get both the data and the mean temperature and write them directly to the CSV file. This is perfectly effective, but it does have a sort of un-Pythonic feel about it, at least to me.
But the bottom line is that with a fairly simple task like this and a very streamlined prompt, Colaboratory as well as Copilot produced code that was usable without changes.
Summary
- Python has the FTP client in the ftplib library to fetch data over the network via FTP.
- The parameiko library can be used to fetch files via SFTP.
- Using a Python script may not be the best choice for fetching files. Be sure to consider options like curl and wget from the command line.
- Using the requests module is your best bet for fetching files by using HTTP/ HTTPS and Python.
- Fetching files from an API using requests is very similar to fetching static files.
- Parameters for API requests often need to be quoted and added as a query string to the request URL.
- The most common structured format used for serving data is JSON.
- XML is a more complex data format for serving data and is still also used.
- Using a library like Beautiful Soup, you can get data by scraping a website and parsing the HTML.
- Scraping sites that you don’t control may not be legal or ethical and requires consideration not to overload the server.
23 Saving data
This chapter covers
- Storing data in relational databases
- Using the Python DB-API
- Accessing databases through an object relational mapper
- Understanding NoSQL databases and how they differ from relational databases
When you have data and have it cleaned, it’s likely that you’ll want to store it. You’ll not only want to store it but also be able to get at it in the future with as little hassle as possible. The need to store and retrieve significant amounts of data usually calls for some sort of database. Relational databases such as PostgreSQL, MySQL, and SQL Server have been established favorites for data storage for decades, and they can still be great options for many use cases. In recent years, NoSQL databases, including MongoDB and Redis (and their various cloud-based relatives), have found favor and can be very useful for a variety of use cases. A detailed discussion of databases would take several books, so in this chapter I look at some scenarios to show how you can access both SQL and NoSQL databases with Python.
23.1 Relational databases
Relational databases have long been a standard for storing and manipulating data. They’re a mature technology and a ubiquitous one. Python can connect with a number of relational databases, but I don’t have the space or the inclination to go through the specifics of each one in this book. Instead, because Python handles databases in a mostly consistent way, I illustrate the basics with one of them—sqlite3—and then discuss some differences and considerations in choosing and using a relational database for data storage.
23.1.1 The Python database API
As I mentioned, Python handles SQL database access very similarly across several database implementations because of PEP-249 (www.python.org/dev/peps/pep-0249/), which specifies some common practices for connecting to SQL databases. Commonly called the Database API or DB-API, it was created to encourage “code that is generally more portable across databases, and a broader reach of database connectivity.” Thanks to the DB-API, the examples of SQLite that you see in this chapter are quite similar to what you’d use for PostgreSQL, MySQL, or several other databases.
23.2 SQLite: Using the sqlite3 database
Although Python has modules for many databases, in the following examples I look at sqlite3. Although it’s not suited for large, high-traffic applications, sqlite3 has two advantages:
- Because it’s part of the standard library, it can be used anywhere you need a database without worrying about adding dependencies.
- sqlite3 stores all of its records in a local file, so it doesn’t need a separate server, which would be the case for PostgreSQL, MySQL, and other larger databases.
These features make sqlite3 a handy option for both smaller applications and quick prototypes.
To use an sqlite3 database, the first thing you need is a Connection object. Getting a Connection object requires calling the connect function with the name of the file that will be used to store the data:
import sqlite3
conn = sqlite3.connect("datafile.db")It’s also possible to hold the data in memory by using “:memory:” as the filename. For storing Python integers, strings, and floats, nothing more is needed. If you want sqlite3 to automatically convert query results for some columns into other types, it’s useful to include the detect_types parameter set to sqlite3.PARSE_DECLTYPES |sqlite3.PARSE_COLNAMES, which directs the Connection object to parse the names and types of columns in queries and attempts to match them with converters you’ve already defined.
The second step is creating a Cursor object from the connection:
cursor = conn.cursor()
cursor
#> <sqlite3.Cursor object at 0xb7a12980>At this point, you’re able to make queries against the database. In the current situation, because the database has no tables or records yet, you first need to create a table and insert a couple of records:
cursor.execute("create table people (id integer primary key, name text, count integer)")
cursor.execute("insert into people (name, count) values ('Bob', 1)")
cursor.execute("insert into people (name, count) values (?, ?)",
("Jill", 15))
conn.commit()The last insert query illustrates the preferred way to make a query with variables. Rather than constructing the query string, it’s more secure to use a ? for each variable and then pass the variables as a tuple parameter to the execute method. The advantage is that you don’t need to worry about incorrectly escaping a value; sqlite3 takes care of it for you.
You can also use variable names prefixed with : in the query and pass in a corresponding dictionary with the values to be inserted:
cursor.execute("insert into people (name, count) values (:username, :usercount)", \
{"username": "Joe", "usercount": 10})
#> <sqlite3.Cursor at 0x7ceda4437ac0>After a table is populated, you can query the data by using SQL commands, again using either ? for variable binding or names and dictionaries:
result = cursor.execute("select * from people")
print(result.fetchall())
#> [(1, 'Bob', 1), (2, 'Jill', 15), (3, 'Joe', 10)]
result = cursor.execute("select * from people where name like :name",
{"name": "bob"})
print(result.fetchall())
#> [(1, 'Bob', 1)]
cursor.execute("update people set count=? where name=?", (20, "Jill"))
result = cursor.execute("select * from people")
print(result.fetchall())
#> [(1, 'Bob', 1), (2, 'Jill', 20), (3, 'Joe', 10)]In addition to the fetchall method, the fetchone method gets one row of the result, and fetchmany returns an arbitrary number of rows. For convenience, it’s also possible to iterate over a cursor object’s rows similarly to iterating over a file:
result = cursor.execute("select * from people")
for row in result:
print(row)
#> (1, 'Bob', 1)
#> (2, 'Jill', 20)
#> (3, 'Joe', 10)Finally, by default, sqlite3 doesn’t immediately commit transactions. This fact means that you have the option of rolling back a transaction if it fails, but it also means that you need to use the Connection object’s commit method to ensure that any changes made have been saved. Doing so before you close a connection to a database is a particularly good idea because the close method doesn’t automatically commit any active transactions:
cursor.execute("update people set count=? where name=?", (20, "Jill"))
conn.commit()
conn.close()Table 23.1 gives an overview of the most common operations on an sqlite3 database.
| Operation | sqlite3 command |
|---|---|
| Create a connection to a database | conn = sqlite3.connect(filename) |
| Create a cursor for a connection | Cursor = conn.cursor() |
| Execute a query with the cursor | cursor.execute(query) |
| Return the results of a query | cursor.fetchall(), cursor.fetchmany(num_rows), cursor.fetchone(), for row in cursor: ... |
| Commit a transaction to a database | conn.commit() |
| Close a connection | conn.close() |
These operations usually are all you need to manipulate an sqlite3 database. Of course, several options let you control their precise behavior; see the Python documentation for more information.
Using sqlite3, write the code that creates a database table for the Illinois weather data you loaded from a flat file in section 22.2. Suppose that you have similar data for more states and want to store more information about the states themselves. How could you modify your database to use a related table to store the state information?
23.3 Using MySQL, PostgreSQL, and other relational databases
As I mentioned earlier in this chapter, several other SQL databases have client libraries that follow the DB-API. As a result, accessing those databases in Python is quite similar, but there are a couple of differences to look out for:
- Unlike SQLite, those databases require a database server that the client connects to and that may or may not be on a different machine, so the connection requires more parameters—usually including host, account name, and password.
- The way in which parameters are interpolated into queries, such as “select * from test where name like :name”, could use a different format—something like ?, %s 5(name)s.
These changes aren’t huge, but they tend to keep code from being completely portable across different databases.
23.4 Making database handling easier with an object relational mapper
There are a few problems with the DB-API database client libraries mentioned earlier in this chapter and their requirement to write raw SQL:
- Different SQL databases have implemented SQL in subtly different ways, so the same SQL statements won’t always work if you move from one database to another, as you might want to do if, say, you do local development against sqlite3 and then want to use MySQL or PostgreSQL in production. Also, as mentioned earlier, the different implementations have different ways of doing things like passing parameters into queries.
- The second drawback is the need to use raw SQL statements. Including SQL statements in your code can make your code more difficult to maintain, particularly if you have a lot of them. In that case, some of the statements will be boilerplate and routine; others will be complex and tricky; and all of them need to be tested, which can get cumbersome.
- The need to write SQL means that you need to think in at least two languages: Python and a specific SQL variant. In plenty of cases, it’s worth these hassles to use raw SQL, but in many other cases, it isn’t.
Given these factors, people wanted a way to handle databases in Python that was easier to manage and didn’t require anything more than writing regular Python code. The solution is an object relational mapper (ORM), which converts, or maps, relational database types and structures to objects in Python. Two of the most common ORMs in the Python world are the Django ORM and SQLAlchemy, although of course there are many others. The Django ORM is rather tightly integrated with the Django web framework and usually isn’t used outside it. Because I’m not delving into Django in this book, I won’t discuss the Django ORM other than to note that it’s the default choice for Django applications and a good one, with fully developed tools and generous community support.
23.4.1 SQLAlchemy
SQLAlchemy is the other big-name ORM in the Python space. SQLAlchemy’s goal is to automate redundant database tasks and provide Python object-based interfaces to the data while still allowing the developer control of the database and access to the underlying SQL. In this section, I look at some basic examples of storing data into a relational database and then retrieving it with SQLAlchemy.
SQLAchemy is already available in Colaboratory, but if you are using a different environment, you can install SQLAlchemy in your environment with pip:
pip install sqlalchemySQLAlchemy offers several ways to interact with the database and its tables. Although an ORM lets you write SQL statements if you want or need to, the strength of an ORM is doing what the name suggests: mapping the relational database tables and columns to Python objects.
We can use SQLAlchemy to replicate what you did in section 23.2: create a table, add three rows, query the table, and update one row. You need to do a bit more setup to use the ORM, but in larger projects, this effort is more than worth it.
First, you import the components you need to connect to the database and map a table to Python objects:
from sqlalchemy import (create_engine, select, MetaData, Table, Column, Integer, String)
from sqlalchemy.orm import sessionmakerFrom the base sqlalchemy package, you need the create_engine and select methods and the MetaData and Table classes. But because you need to specify the schema information when you create the table object, you also need to import the Column class and the classes for the data type of each column—in this case, Integer and String. From the sqlalchemy.orm subpackage, you also need the sessionmaker function.
Now you can think about connecting to the database:
dbPath = 'datafile2.db'
engine = create_engine('sqlite:///%s' % dbPath) # <-- Creates engine object
metadata = MetaData()
people = Table('people', metadata, # <-- Creates people table object
Column('id', Integer, primary_key=True),
Column('name', String),
Column('count', Integer),
)
Session = sessionmaker(bind=engine) # <-- Creates Session class
session = Session()
metadata.create_all(engine) # <-- Creates table in databaseTo create and connect, you need to create a database engine appropriate for your database; then you need a MetaData object, which is a container for managing tables and their schemas. Create a Table object called people, giving the table’s name in the database, the MetaData object you just created, and the column you want to create, as well as their data types. Finally, use the sessionmaker function to create a Session class for your engine and use that class to instantiate a session object. At this point, you’re connected to the database, and the last step is to use the create_all method to create the table.
When the table is created, the next step is inserting some records. Again, you have many options for doing this in SQLAlchemy, but you’ll be fairly explicit in this example. Create an insert object, which you then execute:
people_ins = people.insert().values(name='Bob', count=1)
str(people_ins)
#> 'INSERT INTO people (name, count) VALUES (?, ?)'
session.execute(people_ins)
#> <sqlalchemy.engine.result.ResultProxy object at 0x7f126c6dd438>
session.commit()Here you use the insert() method to create an insert query object, also specifying the fields and values you want to insert. people_ins is the insert object, and you use the str() function to show that, behind the scenes, you created the correct SQL INSERT command. Then you use the session object’s execute method to perform the insertion and the commit method to commit it to the database (remember that you must call commit to have the changes saved to the database):
session.execute(people_ins, [
{'name': 'Jill', 'count':15},
{'name': 'Joe', 'count':10}
])
#> <sqlalchemy.engine.result.ResultProxy object at 0x7f126c6dd908>We could have inserted just one record, but to streamline things a bit we passed in a list of dictionaries of the field names and values for each insert to perform multiple inserts. If we then select the contents of the people table, we can see that we added two new records:
session.commit()
result = session.execute(select(people))
for row in result:
print(row)
#> (1, 'Bob', 1)
#> (2, 'Jill', 15)
#> (3, 'Joe', 10)We can do select operations a bit more directly by instead using the select() method with a where() method to find a particular record:
result = session.execute(select(people).where(people.c.name == 'Jill'))
for row in result:
print(row)
#> (2, 'Jill', 15)In the example, we were looking for any records in which the name column equals ‘Jill’. Note that the where expression uses people.c.name, with the c indicating that name is a column in the people table.
Finally, there is also an update() method we can use to change a value in the database:
result = session.execute(people.update().values(count=20).where(people.c.name == 'Jill'))
session.commit()
result = session.execute(select(people).where(people.c.name == 'Jill'))
for row in result:
print(row)
#> (2, 'Jill', 20)To perform an update, we need to combine the update() method with a values() method that specifies what to update and a where() method to specify the row that update affects.
Mapping table objects to classes
So far, we’ve used table objects directly, but it’s also possible to use SQLAlchemy to map a table directly to a class. This technique has the advantage that the columns are mapped directly to class attributes; for illustration, to make a class People:
from sqlalchemy.orm import declarative_base
Base = declarative_base()
class People(Base):
__tablename__ = "people"
id = Column(Integer, primary_key=True)
name = Column(String)
count = Column(Integer)
results = session.query(People).filter_by(name='Jill')
for person in results:
print(person.id, person.name, person.count)
#> (2, 'Jill', 20)Inserts can be done just by creating an instance of the mapped class and adding it to the session:
new_person = People(name='Jane', count=5)
session.add(new_person)
session.commit()
results = session.query(People).all()
for person in results:
print(person.id, person.name, person.count)
#> 1 Bob 1
#> 2 Jill 20
#> 3 Joe 10
#> 4 Jane 5Updates are also fairly straightforward. You retrieve the record you want to update, change the values on the mapped instance, and then add the updated record to the session to be written back to the database:
jill = session.query(People).filter_by(name='Jill').first()
jill.name
#> 'Jill'
jill.count = 22
session.add(jill)
session.commit()
results = session.query(People).all()
for person in results:
print(person.id, person.name, person.count)
#> 1 Bob 1
#> 2 Jill 22
#> 3 Joe 10
#> 4 Jane 5Deleting is similar to updating; you fetch the record to be deleted and then use the session’s delete() method to delete it:
jane = session.query(People).filter_by(name='Jane').first()
session.delete(jane)
session.commit()
jane = session.query(People).filter_by(name='Jane').first()
print(jane)
NoneUsing SQLAlchemy does take a bit more setup than just using raw SQL, but it also has some real benefits. For one thing, using the ORM means that you don’t need to worry about any subtle differences in the SQL supported by different databases. The example works equally well with sqlite3, MySQL, and PostgreSQL without making any changes in the code other than giving the string to the create_engine and making sure that the correct database driver is available.
Another advantage is that the interaction with the data can happen through Python objects, which may be more accessible to coders who lack SQL experience. Instead of constructing SQL statements, they can use Python objects and methods.
Using the database from earlier, write an SQLAlchemy class to map to the data table and use it to read the records from the table.
23.4.2 Using Alembic for database schema changes
In the course of developing code that uses a relational database, it’s quite common, if not universal, to have to change the structure or schema of the database after you’ve started work. Fields need to be added, or their types need to be changed, and so on. It’s possible, of course, to manually make the changes to both the database tables and to the code for the ORM that accesses them, but that approach has some drawbacks. For one thing, such changes are difficult to roll back if you need to, and it’s hard to keep track of the configuration of the database that goes with a particular version of your code.
The solution is to use a database migration tool to help you make the changes and track them. Migrations are written as code and should include code both to apply the needed changes and to reverse them. Then the changes can be tracked and applied or reversed in the correct sequence. As a result, you can reliably upgrade or downgrade your database to any of the states it was in over the course of development.
As an example, this section looks briefly at Alembic, a popular lightweight migration tool for SQLAlchemy. To start, switch to the system command-line window in your project directory, install Alembic, and create a generic environment by using alemic init:
pip install alembic
alembic init alembicThis code creates the file structure you need to use Alembic for data migrations. There’s an alembic.ini file that you need to edit in at least one place. The squalchemy .url line needs to be updated to match your current situation. In the Colaboratory notebook, there is a cell that will do this if you execute it, but in other environments, you can use a text editor and find the following line:
sqlalchemy.url = driver://user:pass@localhost/dbnameChange the line to
sqlalchemy.url = sqlite:///datafile.dbBecause you’re using a local sqlite file, you don’t need a username or password. The next step is creating a revision by using Alembic’s revision command:
alembic revision -m "create an address table"
#> Generating /home/naomi/qpb_testing/alembic/versions/384ead9efdfd_create_a_
#> test_address_table.py ... done # <-- The revision ID in the filename will be different.This code creates a revision script, 384ead9efdfd_create_a_test_address_table.py, in the alembic/versions directory. This file looks like this:
"""create an address table
Revision ID: 384ead9efdfd # <-- The revision ID in the filename will be different.
Revises:
Create Date: 2017-07-26 21:03:29.042762
"""
from alembic import op
import sqlalchemy as sa
# revision identifiers, used by Alembic.
revision = '384ead9efdfd'
down_revision = None
branch_labels = None
depends_on = None
def upgrade():
pass
def downgrade():
passYou can see that the file contains the revision ID and date in the header. It also contains a down_revision variable to guide the rollback of each version. If you make a second revision, its down_revision variable should contain this revision’s ID.
To perform the revision, we need to update the revision script to supply both the code describing how to perform the revision in the upgrade() method and the code to reverse it in the downgrade() method (this is done in a cell in the notebook):
def upgrade():
op.create_table(
'address',
sa.Column('id', sa.Integer, primary_key=True),
sa.Column('address', sa.String(50), nullable=False),
sa.Column('city', sa.String(50), nullable=False),
sa.Column('state', sa.String(20), nullable=False),
)
def downgrade():
op.drop_table('address')When this code is created, you can apply the upgrade. But first, switch back to the Python shell window to see what tables you have in your database:
for table in metadata.sorted_tables:
print(table.name)
#> peopleAs you might expect, you have only the one table you created earlier. Now you can run Alembic’s upgrade command to apply the upgrade and add a new table. Switch over to your system command line and run
alembic upgrade head
# INFO [alembic.runtime.migration] Context impl SQLiteImpl.
# INFO [alembic.runtime.migration] Will assume non-transactional DDL.
# INFO [alembic.runtime.migration] Running upgrade -> 384ead9efdfd,
# ➥create an address tableIf you pop back to Python and check, you see that the database has two additional tables:
for table in metadata.sorted_tables:
print(table.name)
#> ['alembic_version', 'people', 'address']The first new table, ‘alembic version’, is created by Alembic to help track which version your database is currently on (for reference for future upgrades and downgrades). The second new table, ‘address’, is the table you added through your upgrade and is ready to use.
If you want to roll back the state of the database to what it was before, all you need to do is run Alembic’s downgrade command in the system window. You give the downgrade command -1 to tell Alembic that you want to downgrade by one version:
alembic downgrade -1
# INFO [alembic.runtime.migration] Context impl SQLiteImpl.
# INFO [alembic.runtime.migration] Will assume non-transactional DDL.
# INFO [alembic.runtime.migration] Running downgrade 384ead9efdfd -> , create
# an address tableNow if you check in your Python session, you’ll be back to where you started:
for table in metadata.sorted_tables:
print(table.name)
#> peopleIf you want to, of course, you can run the upgrade again to put the table back, add further revisions, make upgrades, and so on.
Experiment with creating an Alembic upgrade that adds a state table to your database, with columns for ID, state name, and abbreviation. Upgrade and downgrade. What other changes would be needed if you were going to use the state table along with the existing data table?
23.5 NoSQL databases
In spite of their longstanding popularity, relational databases aren’t the only ways to think about storing data. Although relational databases are all about normalizing data within related tables, other approaches look at data differently. Quite commonly, these types of databases are referred to as NoSQL databases, because they usually don’t adhere to the row/column/table structure that SQL was created to describe.
Rather than handle data as collections of rows, columns, and tables, NoSQL databases can look at the data they store as key-value pairs, as indexed documents, and even as graphs. Many NoSQL databases are available, all with somewhat different ways of handling data. In general, they’re less likely to be strictly normalized, which can make retrieving information faster and easier. As examples, in this chapter I look at using Python to access two common NoSQL databases: Redis and MongoDB. What follows barely scratches the surface of what you can do with NoSQL databases and Python, but it should give you a basic idea of the possibilities. If you’re already familiar with Redis or MongoDB, you’ll see a little of how the Python client libraries operate, and if you’re new to NoSQL databases, you’ll at least get an idea of how databases like these work.
Keep in mind that Redis and MongoDB are only two examples of NoSQL databases, chosen to serve as examples. As cloud services have proliferated over the past several years, many other options have arisen, each with particular strengths and weaknesses.
23.6 Key-value stores with Redis
Redis is an in-memory networked key-value store. Because the values are in memory, lookups can be quite fast, and the fact that it’s designed to be accessed over the network makes it useful in a variety of situations. Redis is commonly used for caching, as a message broker, and for quick lookups of information. In fact, the name (which comes from Remote Dictionary Server) is an excellent way to think of it; it behaves much like a Python dictionary translated to a network service.
The following example gives you an idea of how Redis works with Python. If you’re familiar with the Redis command-line interface or have used a Redis client for another language, these short examples should get you well on your way to using Redis with Python. If Redis is new to you, the following gives you an idea of how it works; you can explore more at https://redis.io.
Although several Python clients are available for Redis, at this writing the way to go (according to the Redis website) is one called redis-py. You can install it with pip install redis.
To experiment, you need to have access to a running Redis server. Depending on how much control you want (and how much work you want to do), you have some options.
Perhaps the easiest option for experimentation is to use a cloud-based Redis server hosted on Redis with a free account. This has the advantage that you don’t need to install or maintain a server. A free account is available at https://redis.io/try-free/. This is the recommended option for most readers.
If you have Docker installed, using the Redis Docker instance should also be a quick and easy way to get a server up and running. You should be able to launch a Redis instance from the command line with a command like docker run -p 6379:6379 redis.
On Linux systems, you could also install Redis by using the system package manager, and on Mac systems, brew install redis should work. On Windows systems, you should check the https://redis.io website or search online for the current options for running Redis on Windows. When Redis is installed, you may need to look online for instructions to make sure that the Redis server is running.
Once you are connected to a server, the following are examples of simple Redis interactions with Python. First, you need to import the Redis library and create a Redis connection object:
import redis
r = redis.Redis(
# set your host address below
host='',
port=10032,
# set your password below
password='')You can use several connection options when creating a Redis connection, including the host, port, and password or SSH certificate. If the server is running on localhost on the default port of 6379, no options are needed. When you have the connection, you can use it to access the key-value store.
One of the first things you might do is use the keys() method to get a list of the keys in the database, which returns a list of keys currently stored (if any). Then you can set some keys of different types and try some ways to retrieve their values:
r.keys()
#> [] # <-- Empty because no keys are set yet
r.set('a_key', 'my value')
#> True # <-- Returns True if operation successful
r.keys()
#> [b'a_key']
v = r.get('a_key')
v
#> b'my value'
r.incr('counter')
#> 1
r.get('counter')
#> b'1'
r.incr('counter')
#> 2
r.get('counter')
#> b'2'These examples show how you can get a list of the keys in the Redis database, how to set a key with a value, and how to set a key with a counter variable and increment it. Note that the first time we list the keys we get an empty list, since no keys have been set. When we set a key, we get a return value of True if the operation succeeded. With integer values, we can use the incr method to add 1 to the current value of a key (or start with the value of 1, if the key doesn’t exist).
The following examples deal with storing arrays or lists:
r.rpush("words", "one")
#> 1 # <-- Operation returns new length of array
r.rpush("words", "two")
#> 2 # <-- Operation returns new length of array
r.lrange("words", 0, -1)
#> [b'one', b'two']
r.rpush("words", "three")
#> 3 # <-- Operation returns new length of array
r.lrange("words", 0, -1)
#> [b'one', b'two', b'three']
r.llen("words")
#> 3
r.lpush("words", "zero")
#> 4
r.lrange("words", 0, -1)
#> [b'zero', b'one', b'two', b'three']
r.lrange("words", 2, 2)
#> [b'two']
r.lindex("words", 1)
#> b'one'
r.lindex("words", 2)
#> b'two'When you start the key, “words” isn’t in the database, but the act of adding or pushing a value to the end (from the right, the r in rpush) creates the key, makes an empty list as its value, and then appends the value ‘one’. The return value is 1, the new length of the array. Using rpush again adds another word to the end, and returns the new length, 2. To retrieve the values in the list, you can use the lrange() function, giving the key both a starting index and an ending index, with -1 indicating the end of the list.
Also note that you can add to the beginning, or left side, of the list with lpush(). You can use lindex() to retrieve a single value in the same way as lranger(), except that you give it the index of the value you want.
One feature of Redis that makes it particularly useful for caching is the ability to set an expiration for a key-value pair. After that time has elapsed, the key and value are removed. This technique is particularly useful for using Redis as a cache. You can set the timeout value in seconds when you set the value for a key:
r.setex("timed", 10, "10 seconds")
#> True
r.pttl("timed")
#> 5208
r.pttl("timed")
#> -2
r.pttl("timed")
#> False
b"timed" in r.keys()
#> FalseIn this case, you set the expiration of “timed” by giving the number of seconds (10) as the first parameter after the key. Then, as you use the pttl() method, you can see the time remaining before expiration in milliseconds. When the value expires, the expiration is returned as –2, and both the key and value are automatically removed from the database, shown by checking for b”timed” in r.keys(). This feature and the finegrained control of it that Redis offers are really useful. For simple caches, you may not need to write much more code to have your problem solved.
It’s worth noting that Redis holds its data in memory, so keep in mind that the data isn’t persistent; if the server crashes, some data is likely to be lost. To mitigate the possibility of data loss, Redis has options to manage persistence—everything from writing every change to disk as it occurs to making periodic snapshots at predetermined times to not saving to disk at all. You can also use the Python client’s save() and bgsave() methods to programmatically force a snapshot to be saved, either blocking until the save is complete with save() or saving in the background in the case of bgsave().
In this chapter, I’ve only touched on a small part of what Redis can do, as well as its data types and the ways it can manipulate them. If you’re interested in finding out more, several sources of documentation are available online, including at https://redislabs .com and https://redis-py.readthedocs.io.
What sorts of data and applications would benefit most from a key-value store like Redis?
23.7 Documents in MongoDB
Another popular NoSQL database is MongoDB, which is sometimes called a document-based database because it isn’t organized in rows and columns but instead stores documents. MongoDB is designed to scale across many nodes in multiple clusters while potentially handling billions of documents. In the case of MongoDB, a document is stored in a format called BSON (binary JSON), so a document consists of key-value pairs and looks like a JSON object or Python dictionary. The following examples give you a taste of how you can use Python to interact with MongoDB collections and documents, but a word of warning is appropriate. In situations requiring scale and distribution of data, high insert rates, complex and unstable schemas, and so on, MongoDB is an excellent choice. However, MongoDB isn’t the best choice in many situations, so be sure to investigate your needs and options thoroughly before choosing.
As with Redis, if you want to experiment with MongoDB, you need to have access to a MongoDB server. Again, the simplest and easiest option is to create a free account and instance on MongoDB.com by accessing https://account.mongodb.com/account/register. Once you have created a free account, you can create a free Mongo Atlas cluster for experimentation. Again, this is the recommended option for most readers, but if you don’t feel the need to experiment with MongoDB, you can just read the following section.
Hint: Be sure to save the username, password, and the connection string shown when creating your cluster. If you find that your operations are all timing out, you might need to visit the Network Access option in the Security section of the control panel and “Allow Access from Anywhere,” if simply allowing your current IP address doesn’t work.
As is the case with Redis, if you want to run MongoDB locally, the easiest solution is to run a Docker instance. All you need to do if you have Docker is enter > docker run -p 27017:27017 mongo at the command line. On a Linux system, your package manager should also be able to intall MongoDB locally and the Mac’s brew install mongodb will do it. On Windows systems, check on www.mongodb.com for the Windows version and installation instructions. As with Redis, search online for any instructions on how to configure and start the server.
As is the case with Redis, several Python client libraries connect to MongoDB databases. To give you an idea of how they work, look at pymongo. The first step in using pymongo is installing it, which you can do with pip:
pip install pymongoWhen you have pymongo installed, you can connect to a MongoDB server by creating an instance of MongoClient and specifying the usual connection details:
from pymongo import MongoClient
client = MongoClient(host='*** connection string here ***') # <-- Using the connection string given when you created your instanceWhen you created your cluster, you had to create a username and password, and you were also shown a connection string that used them. If you saved that connection string, you can use it to connect. If you need to find the connection string, in the cluster control panel click the Connect button, then choose the Drivers option; make sure that the Driver is set to Python, and then under step 3, you will see your connection string with your username, but you will need to add the password manually.
MongoDB is organized in terms of a database that contains collections, each of which can contain documents. Databases and collections don’t need to be created before you try to access them, however. If they don’t exist, they’re created as you insert into them, or they simply return no results if you try to retrieve records from them.
To test the client, make a sample document, which can be a Python dictionary:
Here, you connect to a database called my_data and a collection of documents, docs. In this case, they don’t exist, but they’ll be created as you access them. Note that no exceptions were raised even though the database and collection didn’t exist. When you asked for a list of the collections, however, you got an empty list because nothing has been stored in your collection. To store a document, use the collection’s insert() method, which returns the document’s unique ObjectId if the operation is successful:

['docs']Now that you’ve stored a document in the docs collection, it shows up when you ask for the collection names in your database. When the document is stored in the collection, you can query for it, update it, replace it, and delete it:
collection.find_one() # <-- Retrieves first record
# {'_id': ObjectId('66dfb0c090d33ea950f087e1'),
# 'name': 'Jane',
# 'age': 34,
# 'interests': ['Python', 'databases', 'statistics'],
# 'date_added': datetime.datetime(2024, 9, 10, 2, 36, 48, 617000)}The first step is to retrieve the first record with the find_one() method:
from bson.objectid import ObjectId
collection.find_one({"_id":
ObjectId('66dfb0c090d33ea950f087e1')}) # <-- Retrieves record matching specification—in this case, ObjectId
# {'_id': ObjectId('66dfb0c090d33ea950f087e1'),
# 'name': 'Jane',
# 'age': 34,
# 'interests': ['Python', 'databases', 'statistics'],
# 'date_added': datetime.datetime(2024, 9, 10, 2, 36, 48, 617000)}We can also fetch a record by its ObjectId, but to do that we first need to import the ObjectId class from bson.objectid. Using the ObjectId, we can also update a specific record:
collection.update_one({"_id":ObjectId('66dfb0c090d33ea950f087e1')},
{"$set": {"name":"Ann"}}) # <-- Updates record according to contents of $set object
# UpdateResult({'n': 1, 'electionId': ObjectId('7fffffff00000000000002b7'),
# 'opTime': {'ts': Timestamp(1725936314, 21), 't': 695}, 'nModified': 1,
# 'ok': 1.0, '$clusterTime': {'clusterTime': Timestamp(1725936314, 21),
# 'signature': {'hash':
# b'\xd7F\xb8?\xa3a\x87\xd9W\x92\xc4\x17\x7f*\xf8\xbe?\x07\xc2\x07', 'keyId':
# 7368510527980437523}}, 'operationTime': Timestamp(1725936314, 21),
# 'updatedExisting': True}, acknowledged=True)
collection.find_one({"_id":ObjectId('66dfb0c090d33ea950f087e1')})
# {'_id': ObjectId('66dfb0c090d33ea950f087e1'),
# 'name': 'Ann',
# 'age': 34,
# 'interests': ['Python', 'databases', 'statistics'],
# 'date_added': datetime.datetime(2024, 9, 10, 2, 36, 48, 617000)}In updating the record, all fields will be updated to match the object passed in. Similarly, we can replace the record with another object:
collection.replace_one({"_id":ObjectId('66dfb0c090d33ea950f087e1')},
{"name":"Maria"}) # <-- Replaces record with new object
# UpdateResult({'n': 1, 'electionId': ObjectId('7fffffff00000000000002b7'),
# 'opTime': {'ts': Timestamp(1725936426, 6), 't': 695}, 'nModified': 1, 'ok':
# 1.0, '$clusterTime': {'clusterTime': Timestamp(1725936426, 6), 'signature':
# {'hash': b'=\xef,\x1b\xe5\xd5\xa3`\xf9"\xcb\x1a\xb4X\xec\x96\xa8\xbdJN',
# 'keyId': 7368510527980437523}}, 'operationTime': Timestamp(1725936426, 6),
# 'updatedExisting': True}, acknowledged=True)collection.find_one({"_id":ObjectId('66dfb0c090d33ea950f087e1')})
# {'_id': ObjectId('66dfb0c090d33ea950f087e1'),
# 'name': 'Maria'} # <-- The record is replaced with a shorter record.In this case, the entire object has been replaced with another. This may be more efficient, depending on how complex the objects are. Finally, we can delete by ObectId:
collection.delete_one({"_id":ObjectId(
'66dfb0c090d33ea950f087e1')}) # <-- Deletes record matching specification
# DeleteResult({'n': 1, 'electionId': ObjectId('7fffffff00000000000002b7'),
# 'opTime': {'ts': Timestamp(1725936489, 7), 't': 695}, 'ok': 1.0,
# '$clusterTime': {'clusterTime': Timestamp(1725936489, 7), 'signature':
# {'hash':
# b'\xcd\xf3\x18\xdc\xe7\x86\xba\x81\xbdU\\n\xec\xbe\x8e\xc6\xf3\xe8M\xb6',
# 'keyId': 7368510527980437523}}, 'operationTime': Timestamp(1725936489, 7)},
# acknowledged=True)
collection.find_one() # <-- Collection empty, no resultFirst, notice that MongoDB matches according to dictionaries of the fields and their values to match. Dictionaries are also used to indicate operators, such as $lt (less than) and $gt (greater than), as well as commands such as $set for the update. The other thing to notice is that, even though the record has been deleted and the collection is now empty, the collection still exists unless it’s specifically dropped:
db.collection_names()
# ['docs']
collection.drop()
db.collection_names()
#> []MongoDB can do many other things, of course. In addition to operating on one record, versions of the same commands cover multiple records, such as find_many and update_many. MongoDB also supports indexing to improve performance and has several methods to group, count, and aggregate data, as well as a built-in map-reduce method.
Think back over the various data samples you’ve seen so far and other types of data. In your experience, which do you think would be well suited to being stored in a database like MongoDB? Would others clearly not be suited, and if so, why not?
23.8 Creating a database
Choose one of the datasets I’ve discussed in the past few chapters and decide which type of database would be best for storing that data. Create that database and write the code to load the data into it. Then choose the two most common and/or likely types of search criteria and write the code to retrieve both single and multiple matching records. This is an open-ended challenge, so there is no “official” answer. Good luck!
Summary
- Python has a DB-API that provides a generally consistent interface for clients of several relational databases.
- SQLite is a file-based relational database that is part of the Python standard library.
- PostgreSQL and MySQL are popular database servers commonly used with Python.
- Using an ORM can make database code even more standard across databases.
- Using an ORM also lets you access relational databases through Python code and objects rather than SQL queries.
- Tools such as Alembic work with ORMs to use code to make reversible changes to a relational database schema.
- Key-value stores such as Redis provide quick in-memory data access without SQL (NoSQL).
- MongoDB provides scalability and is based on documents, without the strict structure of databases.
24 Exploring data
This chapter covers
- Python’s advantages for handling data
- Using pandas
- Data aggregation
- Plots with Matplotlib
Over the past few chapters, I’ve dealt with some aspects of using Python to get and clean data. Now it’s time to look at a few of the things that Python can help you do to manipulate and explore data.
24.1 Python tools for data exploration
In this chapter, we’ll look at some common Python tools for data exploration in Jupyter: pandas and Matplotlib. I can only touch briefly on a few features of these tools, but the aim is to give you an idea of what is possible and some initial tools to use in exploring data with Python.
24.1.1 Python’s advantages for exploring data
Python has become one of the leading languages for data science and continues to grow in that area. As I’ve mentioned, however, Python isn’t always the fastest language in terms of raw performance. Conversely, some data-crunching libraries, such as NumPy, are largely written in C and heavily optimized to the point that speed isn’t a problem. In addition, considerations such as readability and accessibility often outweigh pure speed; minimizing the amount of developer time needed is often more important. Python is readable and accessible, and both on its own and in combination with tools developed in the Python community, it’s an enormously powerful tool for manipulating and exploring data.
24.1.2 Python can be better than a spreadsheet
Spreadsheets have been the tools of choice for ad hoc data manipulation for decades. People who are skilled with spreadsheets can make them do truly impressive tricks: spreadsheets can combine different but related datasets, pivot tables, use lookup tables to link datasets, and much more. But although people everywhere get a vast amount of work done with them every day, spreadsheets do have limitations, and Python can help you go beyond those limitations.
One limitation that I’ve already alluded to is the fact that most spreadsheet software has a row limit—currently, about 1 million rows, which isn’t enough for many datasets. Another limitation is the central metaphor of the spreadsheet itself. Spreadsheets are two-dimensional grids, rows, and columns or at best stacks of grids, which limits the ways you can manipulate and think about complex data.
With Python, you can code your way around the limitations of spreadsheets and manipulate data the way you want. You can combine Python data structures, such as lists, tuples, sets, and dictionaries, in endlessly flexible ways, or you can create your own classes to package both data and behavior exactly the way you need.
24.2 Python and pandas
In the course of exploring and manipulating data, you perform quite a few common operations, such as loading data into a list or dictionary, cleaning data, and filtering data. Most of these operations are repeated often, have to be done in standard patterns, and are simple and often tedious. If you think that this combination is a strong reason to automate those tasks, you’re not alone. One of the now-standard tools for handling data in Python—pandas—was created to automate the boring heavy lifting of handling datasets.
24.2.1 Why you might want to use pandas
Pandas was created to make manipulating and analyzing tabular or relational data easy by providing a standard framework for holding the data, with convenient tools for frequent operations. As a result, it’s almost more of an extension to Python than a library, and it changes the way you can interact with data. The plus side is that after you grok how pandas work, you can do some impressive things and save a lot of time. It does take time to learn how to get the most from pandas, however. As with many tools, if you use pandas for what it was designed for, it excels. The simple examples I show you in the following sections should give you a rough idea whether pandas is a tool that’s suited for your use cases.
24.2.2 Installing pandas
If you are using Colaboratory, pandas, matplotlib, and numpy are already installed, but if you don’t have pandas, it’s easy to install with pip. It’s often used along with matplotlib for plotting, so you can install both tools from the command line of your Jupyter virtual environment with this code:
pip install pandas matplotlibFrom a cell in a Jupyter notebook, you can use
!pip install pandas matplotlib
If you use pandas, life will be easier if you use the following three lines:
%matplotlib inline
import pandas as pd
import numpy as npThe first line is a Jupyter “magic” function that enables matplotlib to plot data in the cell where your code is (which is very useful). The second line imports pandas with the alias of pd, which is both easier to type and common among pandas users; the last line also imports numpy, which pip will install automatically with pandas. Although pandas depends quite a bit on numpy, you won’t use it explicitly in the following examples, but it’s reasonable to get into the habit of importing it anyway.
24.2.3 Data frames
One basic structure that you get with pandas is a data frame. A data frame is a twodimensional grid, rather similar to a relational database table except in memory. Creating a data frame is easy; you give it some data. To keep things absolutely simple, give it a 3 × 3 grid of numbers as the first example. In Python, such a grid is a list of lists:
grid = [[1,2,3], [4,5,6], [7,8,9]]
print(grid)
#> [[1, 2, 3], [4, 5, 6], [7, 8, 9]]Sadly, in Python, the grid won’t look like a grid unless you make some additional effort. See what you can do with the same grid as a pandas data frame:
import pandas as pd
df = pd.DataFrame(grid)
print(df)
#> 0 1 2
#> 0 1 2 3
#> 1 4 5 6
#> 2 7 8 9That code is fairly straightforward; all you needed to do was turn your grid into a data frame. You’ve gained a more grid-like display, and now you have both row and column numbers. It’s often rather bothersome to keep track of what column number is what, of course, so give your columns names:
df = pd.DataFrame(grid, columns=["one", "two", "three"] )
print(df)
#> one two three
#> 0 1 2 3
#> 1 4 5 6
#> 2 7 8 9You may wonder whether naming the columns has any benefit, but the column names can be put to use with another pandas trick: the ability to select columns by name. If you want the contents only of column “two”, for example, you can get it very simply:
print(df["two"])
#> 0 2
#> 1 5
#> 2 8
#> Name: two, dtype: int64Here, you’ve already saved time in comparison with Python. To get only column 2 of your grid, you’d need to use a list comprehension while also remembering to use a zero-based index (and you still wouldn’t get the nice output):
print([x[1] for x in grid])
#> [2, 5, 8]You can loop over data frame column values just as easily as the list you got by using a comprehension:
for x in df["two"]:
print(x)
#> 2
#> 5
#> 8That’s not bad for a start, but by using a list of columns in double brackets, you can do better, getting a subset of the data frame that’s another data frame. Instead of getting the middle column, get the first and last columns of your data frame as another data frame:
edges = df[["one", "three"]]
print(edges)
#> one three
#> 0 1 3
#> 1 4 6
#> 2 7 9A data frame also has several methods that apply the same operation and argument to every item in the frame. If you want to add 2 to every item in the data frame’s edges, you could use the add() method:
print(edges.add(2))
#> one three
#> 0 3 5
#> 1 6 8
#> 2 9 11Here again, it’s possible to get the same result by using list comprehensions and/or nested loops, but those techniques aren’t as convenient. It’s pretty easy to see how such functionality can make life easier, particularly for someone who’s more interested in the information that the data contains than in the process of cleaning it.
24.3 Data cleaning
In earlier chapters, I discussed a few ways to use Python to clean data. Now that I’ve added pandas to the mix, I’ll show you examples of how to use its functionality to clean data. As I present the following operations, I also refer to ways that the same operation might be done in plain Python, both to illustrate how using pandas is different and to show why pandas isn’t right for every use case (or user, for that matter).
24.3.1 Loading and saving data with pandas
Pandas has an impressive collection of methods to load data from different sources. It supports several file formats (including fixed-width and delimited text files, spreadsheets, JSON, XML, and HTML), but it’s also possible to read from SQL databases, Google BiqQuery, HDF, and even clipboard data. You should be aware that many of these operations aren’t actually part of pandas itself; pandas relies on having other libraries installed to handle those operations, such as SQLAlchemy for reading from SQL databases. This distinction matters mostly if something goes wrong; quite often, the problem that needs to be fixed is outside pandas, and you’re left to deal with the underlying library.
Reading a JSON file with the read_json() method is simple. For example, mars_ data_01.json contains a single day’s weather on Mars as reported by the (now sadly defunct) Opportunity rover. To read it, we can use the pandas read_json method:
mars = pd.read_json("mars_data_01.json")This code gives you a data frame like the following:
report
terrestrial_date 2017-01-11
sol 1576
ls 296.0
min_temp -72.0
min_temp_fahrenheit -97.6
max_temp -1.0
max_temp_fahrenheit 30.2
pressure 869.0
pressure_string Higher
abs_humidity None
wind_speed None
wind_direction --
atmo_opacity Sunny
season Month 10
sunrise 2017-01-11T12:31:00Z
sunset 2017-01-12T00:46:00ZFor another example of how simple reading data into pandas is, we can load some data from the CSV file of temperature data from chapter 22. To read a CSV file, we can use the read_csv() method:
temp = pd.read_csv("temp_data_01.csv")temp = pd.read_csv("temp_data_01.csv", header=0, names=range(18), usecols=range(4,18))
print(temp) 4 5 6 7 8 9 10 11 12 13 14 \
0 1979/01/01 17.48 994 6.0 30.5 2.89 994 -13.6 15.8 Missing 0
1 1979/01/02 4.64 994 -6.4 15.8 -9.03 994 -23.6 6.6 Missing 0
2 1979/01/03 11.05 994 -0.7 24.7 -2.17 994 -18.3 12.9 Missing 0
3 1979/01/04 9.51 994 0.2 27.6 -0.43 994 -16.3 16.3 Missing 0
4 1979/05/15 68.42 994 61.0 75.1 51.30 994 43.3 57.0 Missing 0
5 1979/05/16 70.29 994 63.4 73.5 48.09 994 41.1 53.0 Missing 0
6 1979/05/17 75.34 994 64.0 80.5 50.84 994 44.3 55.7 82.60 2
7 1979/05/18 79.13 994 75.5 82.1 55.68 994 50.0 61.1 81.42 349
8 1979/05/19 74.94 994 66.9 83.1 58.59 994 50.9 63.2 82.87 78
15 16 17
0 Missing Missing 0.00%
1 Missing Missing 0.00%
2 Missing Missing 0.00%
3 Missing Missing 0.00%
4 Missing Missing 0.00%
5 Missing Missing 0.00%
6 82.40 82.80 0.20%
7 80.20 83.40 35.11%
8 81.60 85.20 7.85% Clearly, loading the file in a single step is appealing, and you can see that pandas had no problems loading the file. You can also see that the empty first column has been translated into NaN (not a number). You do still have the same problem with ‘Missing’ for some values, and in fact it might make sense to have those ‘Missing’ values converted to NaN:
temp = pd.read_csv("temp_data_01.csv", na_values=['Missing'])The addition of the na_values parameter controls what values will be translated to NaN on load. In this case, you added the string ‘Missing’ so that the row of the data frame was translated from
NaN Illinois 17 Jan 01, 1979 1979/01/01 17.48 994 6.0 30.5
➥2.89 994 -13.6 15.8 Missing 0 Missing Missing 0.00%to
NaN Illinois 17 Jan 01, 1979 1979/01/01 17.48 994 6.0 30.5
➥2.89 994 -13.6 15.8 NaN 0 NaN NaN 0.00%This technique can be particularly useful if you have one of those data files in which, for whatever reason, “no data” is indicated in a variety of ways: NA, N/A, ?, -, and so on. To handle a case like that, you can inspect the data to find out what’s used and then reload it using the na_values parameter to standardize all those variations as NaN.
Saving data
If you want to save the contents of a data frame, a pandas data frame has a similarly broad collection of methods. If you take your simple grid data frame, you can write it in several ways. The following line
df.to_csv("df_out.csv", index=False) # <-- Setting index to False means that the row indexes will not be written.writes a file that looks like this:
one,two,three
1,2,3
4,5,6
7,8,9Similarly, you can transform a data grid to a JSON object or write it to a file:
df.to_json() # <-- Supplying a file path as an argument writes the JSON to that file rather than returning it.
#> '{"one":{"0":1,"1":4,"2":7},"two":{"0":2,"1":5,"2":8},
#> "three":{"0":3,"1":6,"2":9}}' 24.3.2 Data cleaning with a data frame
Converting a particular set of values to NaN on load is a very simple bit of data cleaning that pandas makes trivial. Going beyond that, data frames support several operations that can make data cleaning less of a chore. To see how this works, reopen the temperature CSV file, but this time, instead of using the headers to name the columns, use the range() function with the names parameter to give them numbers, which will make referring to them easier. You also may recall from an earlier example that the first field of every line—the “Notes” field—is empty and loaded with NaN values. Although you could ignore this column, it would be even easier if you didn’t have it. You can use the range() function again, this time starting from 1, to tell pandas to load all columns except the first one. But if you know that all of your values are from Illinois and you don’t care about the long-form date field, you could start from 4 to make things much more manageable:
temp = pd.read_csv("temp_data_01.csv", na_values=['Missing'], header=0,
names=range(18), usecols=range(4,18)) # <-- Setting header = 0 turns off reading the header for column labels.
print(temp) 4 5 6 7 8 9 10 11 12 13 14 \
0 1979/01/01 17.48 994 6.0 30.5 2.89 994 -13.6 15.8 NaN 0
1 1979/01/02 4.64 994 -6.4 15.8 -9.03 994 -23.6 6.6 NaN 0
2 1979/01/03 11.05 994 -0.7 24.7 -2.17 994 -18.3 12.9 NaN 0
3 1979/01/04 9.51 994 0.2 27.6 -0.43 994 -16.3 16.3 NaN 0
4 1979/05/15 68.42 994 61.0 75.1 51.30 994 43.3 57.0 NaN 0
5 1979/05/16 70.29 994 63.4 73.5 48.09 994 41.1 53.0 NaN 0
6 1979/05/17 75.34 994 64.0 80.5 50.84 994 44.3 55.7 82.60 2
7 1979/05/18 79.13 994 75.5 82.1 55.68 994 50.0 61.1 81.42 349
8 1979/05/19 74.94 994 66.9 83.1 58.59 994 50.9 63.2 82.87 78
15 16 17
0 NaN NaN 0.00%
1 NaN NaN 0.00%
2 NaN NaN 0.00%
3 NaN NaN 0.00%
4 NaN NaN 0.00%
5 NaN NaN 0.00%
6 82.4 82.8 0.20%
7 80.2 83.4 35.11%
8 81.6 85.2 7.85% Now you have a data frame that has only the columns you might want to work with. But you still have a problem: the last column, which lists the percentage of coverage for the heat index, is still a string ending with a percentage sign rather than an actual percentage. This problem is apparent if you look at the first row’s value for column 17:
temp[17][0]
#> '0.00%'To fix this problem, you need to do two things: remove the % from the end of the value and then cast the value from a string to a number. Optionally, if you want to represent the resulting percentage as a fraction, divide it by 100. The first bit is simple because pandas lets you use a single command to repeat an operation on a column:
temp[17] = temp[17].str.strip("%")
temp[17][0]
#> '0.00'This code takes the column and calls a string strip() operation on it to remove the trailing %. Now when you look at the first value in the column (or any of the other values), you see that the offending percentage sign is gone. It’s also worth noting that you could have used other operations, such as replace(“%”, ““), to achieve the same result.
The second operation is to convert the string to a numeric value. Again, pandas lets you perform this operation with one command:
temp[17] = pd.to_numeric(temp[17])
temp[17][0]
#> 0.0Now the values in column 17 are numeric, and if you want to, you can use the div() method to finish the job of turning those values into fractions:
temp[17] = temp[17].div(100)
temp[17]
#> 0 0.0000
#> 1 0.0000
#> 2 0.0000
#> 3 0.0000
#> 4 0.0000
#> 5 0.0000
#> 6 0.0020
#> 7 0.3511
#> 8 0.0785
#> Name: 17, dtype: float64In fact, it would be possible to achieve the same result in a single line by chaining the three operations together:
temp[17] = pd.to_numeric(temp[17].str.strip("%")).div(100)This example is very simple, but it gives you an idea of the convenience that pandas can bring to cleaning your data. Pandas has a wide variety of operations for transforming data, as well as the ability to use custom functions, so it would be hard to think of a scenario in which you couldn’t streamline data cleaning with pandas.
Although the number of options is almost overwhelming, a wide variety of tutorials and videos is available in the documentation at http://pandas.pydata.org excellent.
Experiment with the operations. When the final column has been converted to a fraction, can you think of a way to convert it back to a string with the trailing percentage sign?
By contrast, load the same data into a plain Python list by using the csv module, and apply the same changes by using plain Python.
24.4 Data aggregation and manipulation
The preceding examples provide some idea of the many options pandas gives you for performing fairly complex operations on your data with only a few commands. As you might expect, this level of functionality is also available for aggregating data. In this section, I walk through a few simple examples of aggregating data to illustrate some of the many possibilities. Although many options are available, I focus on merging data frames, performing simple data aggregation, and grouping and filtering.
24.4.1 Merging data frames
Quite often in the course of handling data, you need to relate two datasets. Suppose that you have one file containing the number of sales calls made per month by members of a sales team and another file that gives the dollar amounts of the sales in each of their territories:
calls = pd.read_csv("sales_calls.csv")
print(calls) Team member Territory Month Calls
0 Jorge 3 1 107
1 Jorge 3 2 88
2 Jorge 3 3 84
3 Jorge 3 4 113
4 Ana 1 1 91
5 Ana 1 2 129
6 Ana 1 3 96
7 Ana 1 4 128
8 Ali 2 1 120
9 Ali 2 2 85
10 Ali 2 3 87
11 Ali 2 4 87revenue = pd.read_csv("sales_revenue.csv")
print(revenue) Territory Month Amount
0 1 1 54228
1 1 2 61640
2 1 3 43491
3 1 4 52173
4 2 1 36061
5 2 2 44957
6 2 3 35058
7 2 4 33855
8 3 1 50876
9 3 2 57682
10 3 3 53689
11 3 4 49173Clearly, it would be very useful to link revenue and team member activity. These two files are very simple, yet merging them with plain Python isn’t entirely trivial. Pandas has a function to merge two data frames:
calls_revenue = pd.merge(calls, revenue, on=['Territory', 'Month'])The merge function creates a new data frame by joining the two frames on the columns specified in the column field. The merge function works similarly to a relational database join, giving you a table that combines the columns from the two files:
print(calls_revenue) Team member Territory Month Calls Amount
0 Jorge 3 1 107 50876
1 Jorge 3 2 88 57682
2 Jorge 3 3 84 53689
3 Jorge 3 4 113 49173
4 Ana 1 1 91 54228
5 Ana 1 2 129 61640
6 Ana 1 3 96 43491
7 Ana 1 4 128 52173
8 Ali 2 1 120 36061
9 Ali 2 2 85 44957
10 Ali 2 3 87 35058
11 Ali 2 4 87 33855In this case, you have a one-to-one correspondence between the rows in the two fields, but the merge function can also do one-to-many and many-to-many joins, as well as right and left joins.
How would you go about merging two datasets like the ones in the Python example?
24.4.2 Selecting data
It can also be useful to select or filter the rows in a data frame based on some condition. In the example sales data, you may want to look only at territory 3, which is also easy:
print(calls_revenue[calls_revenue.Territory==3]) Team member Territory Month Calls Amount
0 Jorge 3 1 107 50876
1 Jorge 3 2 88 57682
2 Jorge 3 3 84 53689
3 Jorge 3 4 113 49173In this example, you select only rows in which the territory is equal to 3 but using exactly that expression, revenue.Territory==3, as the index for the data frame. From the point of view of plain Python, such use is nonsense and illegal, but for a pandas data frame, it works and makes for a much more concise expression.
More complex expressions are also allowed, of course. If you want to select only rows in which the amount per call is greater than 500, you could use this expression instead:
print(calls_revenue[calls_revenue.Amount/calls_revenue.Calls>500]) Team member Territory Month Calls Amount
1 Jorge 3 2 88 57682
2 Jorge 3 3 84 53689
4 Ana 1 1 91 54228
9 Ali 2 2 85 44957Even better, you could calculate and add that column to your data frame by using a similar operation:
calls_revenue['Call_Amount'] = calls_revenue.Amount/calls_revenue.Calls
print(calls_revenue) Team member Territory Month Calls Amount Call_Amount
0 Jorge 3 1 107 50876 475.476636
1 Jorge 3 2 88 57682 655.477273
2 Jorge 3 3 84 53689 639.154762
3 Jorge 3 4 113 49173 435.159292
4 Ana 1 1 91 54228 595.912088
5 Ana 1 2 129 61640 477.829457
6 Ana 1 3 96 43491 453.031250
7 Ana 1 4 128 52173 407.601562
8 Ali 2 1 120 36061 300.508333
9 Ali 2 2 85 44957 528.905882
10 Ali 2 3 87 35058 402.965517
11 Ali 2 4 87 33855 389.137931Again, note that pandas’ built-in logic replaces a more cumbersome structure in plain Python.
What Python code structure would you use to select only rows meeting certain conditions?
24.4.3 Grouping and aggregation
As you might expect, pandas has plenty of tools to summarize and aggregate data as well. In particular, getting the sum, mean, median, minimum, and maximum values from a column uses clearly named column methods:
print(calls_revenue.Calls.sum())
print(calls_revenue.Calls.mean())
print(calls_revenue.Calls.median())
print(calls_revenue.Calls.max())
print(calls_revenue.Calls.min())
#> 1215
#> 101.25
#> 93.5
#> 129
#> 84If, for example, you want to get all of the rows in which the amount per call is above the median, you can combine this trick with the selection operation:
print(calls_revenue.Call_Amount.median())
print(calls_revenue[calls_revenue.Call_Amount >= calls_revenue.Call_Amount.median()])464.2539427570093
Team member Territory Month Calls Amount Call_Amount
0 Jorge 3 1 107 50876 475.476636
1 Jorge 3 2 88 57682 655.477273
2 Jorge 3 3 84 53689 639.154762
4 Ana 1 1 91 54228 595.912088
5 Ana 1 2 129 61640 477.829457
9 Ali 2 2 85 44957 528.905882In addition to being able to pick out summary values, it’s often useful to group the data based on other columns. In this simple example, you can use the groupby method to group your data. You may want to know the total calls and amounts by month or by territory, for example. In those cases, use those fields with the data frame’s groupby method:
print(calls_revenue[['Month', 'Calls', 'Amount']].groupby(['Month']).sum()) Calls Amount
Month
1 318 141165
2 302 164279
3 267 132238
4 328 135201print(calls_revenue[['Territory', 'Calls', 'Amount']].groupby(['Territory']). sum()) Calls Amount
Territory
1 444 211532
2 379 149931
3 392 211420
In each case, you select the columns that you want to aggregate, group them by the values in one of those columns, and (in this case) sum the values for each group. You could also use any of the other methods mentioned earlier in this chapter.
Again, all these examples are simple, but they illustrate a few of the options you have for manipulating and selecting data with pandas. If these ideas resonate with your needs, you can learn more by studying the pandas documentation: https://pandas.pydata.org/.
Experiment with pandas and the data in previous examples. Can you get the calls and amounts by both team member and month?
24.5 Plotting data
Another very attractive feature of pandas is the ability to plot the data in a data frame very easily. Although you have many options for plotting data in Python and Jupyter notebooks, pandas can use matplotlib directly from a data frame. (While matplotlib has a rich set of plotting and graphics functions that are worth exploration, here we’ll just touch on some simple examples. If you are interested in data visualization, you will want to explore matplotlib more deeply.) You may recall that when you started your Jupyter session, one of the first commands you gave was the Jupyter “magic” command to enable matplotlib for inline plotting:
%matplotlib inlineBecause you have the ability to plot, see how you might plot some data (figure 24.1). To continue with the sales example, if you want to plot the quarter’s mean sales by territory, you can get a graph right in your notebook just by adding .plot.bar():
calls_revenue[['Territory', 'Calls']].groupby(['Territory']).sum().plot.bar()Other options are available. plot() alone and .plot.line() create a line graph, .plot.pie() creates a pie chart, and so on.
Thanks to the combination of pandas and matplotlib, plotting data in a Jupyter notebook is quite easy. I should also note that, although such plotting is easy, there are many things that this combination doesn’t do extremely well.

Figure 24.1 Bar plot of a pandas data frame in Jupyter notebook
Plot a line graph of the monthly average amount per call.
24.6 Why you might not want to use pandas
The preceding examples illustrate only a tiny fraction of the tools pandas can offer you in cleaning, exploring, and manipulating data. As I mentioned at the beginning of this chapter, pandas is an excellent tool set that shines in what it was designed to do. That doesn’t mean, however, that pandas is the tool for all situations or for all people.
There are reasons why you might elect to use plain old Python (or some other tool) instead. For one thing, as I mentioned earlier, learning to fully use pandas is in some ways like learning another language, which may not be something you have the time or inclination to do. Also, pandas may not be ideal in all production situations, particularly with very large datasets that don’t require much in the way of math operations or with data that isn’t easy to put into the formats that work best with pandas. Munging large collections of product information, for example, probably wouldn’t benefit so much from pandas; neither would basic processing of a stream of transactions.
The point is that you should choose your tools thoughtfully based on the problems at hand. In many cases, pandas will truly make your life easier as you work with data, but in other cases, plain old Python may be your best bet.
Summary
Python offers many benefits for data handling, including the ability to handle very large datasets and the flexibility to handle data in ways that match your needs.
- By allowing you to select, group, and otherwise manipulate data elements, Python often gives you more powerful and flexible data operations than a spreadsheet.
- Pandas is a tool that makes many common data-handling operations much easier.
- A data frame is the structure pandas uses to manipulate data.
- Pandas has methods for loading and saving data from files in several common data formats, including CSV and JSON.
- Pandas operations on columns makes data cleaning easier, operating by column on all rows of the data.
- The pandas merge method allows you to combine data frames on a shared field.
- Pandas combined with matplotlib also makes simple plotting easy by allowing you to plot data with a single plot command.
- In spite of its many advantages, pandas may not always be the best choice, particularly for large datasets that aren’t suited to pandas or need near-real-time processing.
Case study
In this case study, you walk through using Python to fetch some data, clean it, and then graph it. This project may be a short one, but it combines several features of the language discussed in this book, and it gives you a chance to see a project worked through from beginning to end. At almost every step, I briefly call out alternatives and enhancements that you can make.
Global temperature change is the topic of much discussion, but those discussions are based on a global scale. Suppose that you want to know what the temperatures have been doing near where you are. One way of finding out is to get historical data for your location, process that data, and plot it to see exactly what’s been happening.
The following case study was done by using a Jupyter notebook in Colaboratory, the same as the code in the other chapters. You can find the notebook I used (with this text and code) in the source code repository at https://github.com/nceder/qpb4e in the code/Case Study folder as Case_Study.ipynb. You can also execute the code in a standard Python shell, and a version that supports that shell is in the source repository as Case Study.py. Note, however, that you will have to install the packages for requests, pandas, and matplotlib before running the code.
Fortunately, several sources of historical weather data are freely available. I’m going to walk you through using data from the Global Historical Climatology Network, which has data from around the world. You may find other sources, which may have different data formats, but the steps and the processes I discuss here should be generally applicable to any dataset.
Downloading the data
The first step will be to get the data. An archive of daily historical weather data (https://www1.ncdc.noaa.gov/pub/data/ghcn/daily/) has a wide array of data. The first step is to figure out which files you want and exactly where they are; then you download them. When you have the data, you can move on to processing and ultimately displaying your results.
To download the files, which are accessible via HTTPS, you need the requests library, which is already available in Colaboratory. If you are using another environment, you can get requests with pip install requests at the command prompt. When you have requests, your first step is to fetch the readme.txt file, which can guide you as to the formats and location of the data files you want:
# import requests
import requests
# get readme.txt file
r = requests.get('https://www1.ncdc.noaa.gov/pub/data/ghcn/daily/readme.txt')
readme = r.textWhen you look at the readme file, you should see something like this:
print(readme)README FILE FOR DAILY GLOBAL HISTORICAL CLIMATOLOGY NETWORK (GHCN-DAILY)
Version 3.22
----------------------------------------------------------------------------
How to cite:
Note that the GHCN-Daily dataset itself now has a DOI (Digital Object
Identifier)
so it may be relevant to cite both the methods/overview journal article as
➥well
as the specific version of the dataset used.
The journal article describing GHCN-Daily is:
Menne, M.J., I. Durre, R.S. Vose, B.E. Gleason, and T.G. Houston, 2012: An
➥overview
of the Global Historical Climatology Network-Daily Database. Journal of
➥Atmospheric
and Oceanic Technology, 29, 897-910, doi:10.1175/JTECH-D-11-00103.1.
To acknowledge the specific version of the dataset used, please cite:
Menne, M.J., I. Durre, B. Korzeniewski, S. McNeal, K. Thomas, X. Yin, S.
➥Anthony, R. Ray,
R.S. Vose, B.E.Gleason, and T.G. Houston, 2012: Global Historical
➥Climatology Network -
Daily (GHCN-Daily), Version 3. [indicate subset used following decimal,
e.g. Version 3.12].
NOAA National Climatic Data Center. http://doi.org/10.7289/V5D21VHZ [access
➥date].In particular, you’re interested in section II, which lists the contents:
II. CONTENTS OF ftp://ftp.ncdc.noaa.gov/pub/data/ghcn/daily
all: Directory with ".dly" files for all of GHCN-Daily
gsn: Directory with ".dly" files for the GCOS Surface Network
(GSN)
hcn: Directory with ".dly" files for U.S. HCN
by_year: Directory with GHCN Daily files parsed into yearly
subsets with observation times where available. See the
/by_year/readme.txt and
/by_year/ghcn-daily-by_year-format.rtf
files for further information
grid: Directory with the GHCN-Daily gridded dataset known
as HadGHCND
papers: Directory with pdf versions of journal articles relevant
to the GHCN-Daily dataset
figures: Directory containing figures that summarize the inventory
of GHCN-Daily station records
ghcnd-all.tar.gz: TAR file of the GZIP-compressed files in the "all"
➥directory
ghcnd-gsn.tar.gz: TAR file of the GZIP-compressed "gsn" directory
ghcnd-hcn.tar.gz: TAR file of the GZIP-compressed "hcn" directory
ghcnd-countries.txt: List of country codes (FIPS) and names
ghcnd-inventory.txt: File listing the periods of record for each station and
element
ghcnd-stations.txt: List of stations and their metadata (e.g.,
➥coordinates)
ghcnd-states.txt: List of U.S. state and Canadian Province codes
used in ghcnd-stations.txt
ghcnd-version.txt: File that specifies the current version of GHCN Daily
readme.txt: This file
status.txt: Notes on the current status of GHCN-DailyAs you look at the files available, you see that ghcnd-inventory.txt has a listing of the recording periods for each station, which will help you find a good dataset, and ghcnd-stations.txt lists the stations, which should help you find the station closest to your location, so you’ll grab those two files first:
II. CONTENTS OF ftp://ftp.ncdc.noaa.gov/pub/data/ghcn/daily
all: Directory with ".dly" files for all of GHCN-Daily
gsn: Directory with ".dly" files for the GCOS Surface Network
(GSN)
hcn: Directory with ".dly" files for U.S. HCN
by_year: Directory with GHCN Daily files parsed into yearly
subsets with observation times where available. See
the
/by_year/readme.txt and
/by_year/ghcn-daily-by_year-format.rtf
files for further information
grid: Directory with the GHCN-Daily gridded dataset known
as HadGHCND
papers: Directory with pdf versions of journal articles relevant
to the GHCN-Daily dataset
figures: Directory containing figures that summarize the inventory
of GHCN-Daily station records
ghcnd-all.tar.gz: TAR file of the GZIP-compressed files in the "all"
directory
ghcnd-gsn.tar.gz: TAR file of the GZIP-compressed "gsn" directory
ghcnd-hcn.tar.gz: TAR file of the GZIP-compressed "hcn" directory
ghcnd-countries.txt: List of country codes (FIPS) and names
ghcnd-inventory.txt: File listing the periods of record for each station and
element
ghcnd-stations.txt: List of stations and their metadata (e.g.,
➥coordinates)
ghcnd-states.txt: List of U.S. state and Canadian Province codes
used in ghcnd-stations.txt
ghcnd-version.txt: File that specifies the current version of GHCN Daily
readme.txt: This file
status.txt: Notes on the current status of GHCN-Daily
# get inventory and stations files
r = requests.get('https://www1.ncdc.noaa.gov/pub/data/ghcn/daily/ghcnd-
➥inventory.txt')
inventory_txt = r.text
r = requests.get('https://www1.ncdc.noaa.gov/pub/data/ghcn/daily/ghcnd-
➥stations.txt')
stations_txt = r.textWhen you have those files, you can save them to your local disk so that you won’t need to download them again if you need to go back to the original data:
# save both the inventory and stations files to disk, in case we need them
with open("inventory.txt", "w") as inventory_file:
inventory_file.write(inventory_txt)
with open("stations.txt", "w") as stations_file:
stations_file.write(stations_txt)Start by looking at the inventory file. Here’s what the first 137 characters show you:
print(inventory\_txt[:137])
#> ACW00011604 17.1167 -61.7833 TMAX 1949 1949
#> ACW00011604 17.1167 -61.7833 TMIN 1949 1949
#> ACW00011604 17.1167 -61.7833 PRCP 1949 1949If we look at section VII of the readme.txt file, we can see that the format of the inventory file is
VII. FORMAT OF "ghcnd-inventory.txt"
------------------------------
Variable Columns Type
------------------------------
ID 1-11 Character
LATITUDE 13-20 Real
LONGITUDE 22-30 Real
ELEMENT 32-35 Character
FIRSTYEAR 37-40 Integer
LASTYEAR 42-45 Integer
------------------------------These variables have the following definitions:
ID is the station identification code. Please see "ghcnd-
➥stations.txt"
for a complete list of stations and their metadata.
LATITUDE is the latitude of the station (in decimal degrees).
LONGITUDE is the longitude of the station (in decimal degrees).
ELEMENT is the element type. See section III for a definition of
➥elements.
FIRSTYEAR is the first year of unflagged data for the given element.
LASTYEAR is the last year of unflagged data for the given element.From this description, you can tell that the inventory list has most of the information you need to find the station you want to look at. You can use the latitude and longitude to find the stations closest to you; then you can use the FIRSTYEAR and LASTYEAR fields to find a station with records covering a long span of time.
The only question remaining is what the ELEMENT field is; for that, the file suggests that you look at section III. In section III (which I look at in more detail later), you find the following description of the main elements:
ELEMENT is the element type. There are five core elements as well as a
number of addition elements.
The five core elements are:
PRCP = Precipitation (tenths of mm)
SNOW = Snowfall (mm)
SNWD = Snow depth (mm)
TMAX = Maximum temperature (tenths of degrees C)
TMIN = Minimum temperature (tenths of degrees C)For purposes of this example, you’re interested in the TMAX and TMIN elements, which are maximum and minimum temperatures in tenths of degrees Celsius.
Parsing the inventory data
The readme.txt file tells you what you’ve got in the inventory file so that you can parse the data into a more usable format. You could just store the parsed inventory data as a list of lists or list of tuples, but it takes only a little more effort to use dataclass from the dataclasses library to create a custom class with the attributes named (a named tuple would also be a possibility here):
# use dataclass to create a custom Inventory class
from dataclasses import dataclass
@dataclass
class Inventory:
station:str
latitude: float
longitude: float
element:str
start:int
end:intUsing the Inventory class you created is very straightforward; you simply create each instance from the appropriate values, which in this case are a parsed row of inventory data.
The parsing involves two steps. First, you need to pick out slices of a line according to the field sizes specified. As you look at the field descriptions in the readme file, it’s also clear that there’s an extra space between files, which you need to consider in coming up with any approach to parsing. In this case, because you’re specifying each slice, the extra spaces are ignored. In addition, because the sizes of the STATION and ELEMENT fields exactly correspond to the values stored in them, you shouldn’t need to worry about stripping excess spaces from them.
The second thing that would be nice to do is convert the latitude and longitude values to floats and the start and end years to ints. You could do this at a later stage of data cleaning, and in fact, if the data is inconsistent and doesn’t have values that convert correctly in every row, you might want to wait. But in this case, the data lets you handle these conversions in the parsing step, so do it now:
# parse inventory lines and convert some values to floats and ints
inventory = [Inventory(x[0:11], float(x[12:20]), float(x[21:30]), x[31:35],
int(x[36:40]), int(x[41:45]))
for x in inventory_txt.split("\n") if x.startswith("US")]
for line in inventory[:5]:
print(line)Inventory(station='US009052008', latitude=43.7333, longitude=-96.6333,
element='TMAX', start=2008, end=2016)
Inventory(station='US009052008', latitude=43.7333, longitude=-96.6333,
element='TMIN', start=2008, end=2016)
Inventory(station='US009052008', latitude=43.7333, longitude=-96.6333,
element='PRCP', start=2008, end=2016)
Inventory(station='US009052008', latitude=43.7333, longitude=-96.6333,
element='SNWD', start=2009, end=2016)
Inventory(station='US10RMHS145', latitude=40.5268, longitude=-105.1113,
element='PRCP', start=2004, end=2004)Selecting a station based on latitude and longitude
Now that the inventory is loaded, you can use the latitude and longitude to find the stations closest to your location and then pick the one with the longest run of temperatures based on start and end years. At even the first line of the data, you can see two things to worry about:
- There are various element types, but you’re concerned only with TMIN and TMAX, for minimum and maximum temperature.
- None of the first inventory entries you see covers more than a few years. If you’re going to be looking for a historical perspective, you want to find a much longer run of temperature data.
To pick out what you need quickly, you can use a list comprehension to make a sublist of only the station inventory items in which the element is TMIN or TMAX. The other thing that you care about is getting a station with a long run of data, so while you’re creating this sublist, also make sure that the start year is before 1920 and that the end year is at least 2024. That way, you’re looking only at stations with over 100 years’ worth of data:
inventory_temps = [x for x in inventory if x.element in ['TMIN', 'TMAX']
and x.end >= 2015 and x.start < 1920]
inventory_temps[:5][Inventory(station='USC00010583', latitude=30.8839, longitude=-87.7853,
element='TMAX', start=1915, end=2024),
Inventory(station='USC00010583', latitude=30.8839, longitude=-87.7853,
element='TMIN', start=1915, end=2024),
Inventory(station='USC00011694', latitude=32.8158, longitude=-86.6044,
element='TMAX', start=1893, end=2024),
Inventory(station='USC00011694', latitude=32.8158, longitude=-86.6044,
element='TMIN', start=1893, end=2024),
Inventory(station='USC00012813', latitude=30.5467, longitude=-87.8808,
element='TMAX', start=1917, end=2024)]Looking at the first five records in your new list, you see that you’re in better shape. Now you have only temperature records, and the start and end years show that you have longer runs.
That leaves the problem of selecting the station nearest your location. To do that, compare the latitude and longitude of the station inventories with those of your location. There are various ways to get the latitude and longitude of any place, but probably the easiest way is to use an online mapping application or online search. (When I do that for the Chicago Loop, I get a latitude of 41.882 and a longitude of -87.629.)
Because you’re interested in the stations closest to your location, that interest implies sorting based on how close the latitude and longitude of the stations are to those of your location. Sorting a list is easy enough, and sorting by latitude and longitude isn’t too hard. But how do you sort by the distance from your latitude and longitude?
The answer is to define a key function for your sort that gets the difference between your latitude and the station’s latitude and the difference between your longitude and the station’s longitude and combines them into one number. The only other thing to remember is that you’ll want to add the absolute value of the differences before you combine them to avoid having a high negative difference combined with an equally high positive difference that would fool your sort:
# Downtown Chicago, obtained via online map
latitude, longitude = 41.882, -87.629
inventory_temps.sort(key=lambda x: abs(latitude-x.latitude) +
abs(longitude-x.longitude))
inventory_temps[:20][Inventory(station='USC00110338', latitude=41.7803, longitude=-88.3092,
element='TMAX', start=1893, end=2024),
Inventory(station='USC00110338', latitude=41.7803, longitude=-88.3092,
element='TMIN', start=1893, end=2024),
Inventory(station='USC00112736', latitude=42.0628, longitude=-88.2861,
element='TMAX', start=1897, end=2024),
Inventory(station='USC00112736', latitude=42.0628, longitude=-88.2861,
element='TMIN', start=1897, end=2024),
Inventory(station='USC00476922', latitude=42.7028, longitude=-87.7858,
element='TMAX', start=1896, end=2024),
Inventory(station='USC00476922', latitude=42.7028, longitude=-87.7858,
element='TMIN', start=1896, end=2024),
Inventory(station='USC00124837', latitude=41.6117, longitude=-86.7297,
element='TMAX', start=1897, end=2024),
Inventory(station='USC00124837', latitude=41.6117, longitude=-86.7297,
element='TMIN', start=1897, end=2024),
Inventory(station='USC00115825', latitude=41.3714, longitude=-88.4333,
element='TMAX', start=1912, end=2024),
Inventory(station='USC00115825', latitude=41.3714, longitude=-88.4333,
element='TMIN', start=1912, end=2024),
Inventory(station='USC00200710', latitude=42.1244, longitude=-86.4267,
element='TMAX', start=1893, end=2024),
Inventory(station='USC00200710', latitude=42.1244, longitude=-86.4267,
element='TMIN', start=1893, end=2024),
Inventory(station='USC00114198', latitude=40.4664, longitude=-87.685,
element='TMAX', start=1902, end=2024),
Inventory(station='USC00114198', latitude=40.4664, longitude=-87.685,
element='TMIN', start=1902, end=2024),
Inventory(station='USW00014848', latitude=41.7072, longitude=-86.3164,
element='TMAX', start=1893, end=2024),
Inventory(station='USW00014848', latitude=41.7072, longitude=-86.3164,
element='TMIN', start=1893, end=2024),
Inventory(station='USC00124657', latitude=41.3053, longitude=-86.6286,
element='TMAX', start=1897, end=2024),
Inventory(station='USC00124657', latitude=41.3053, longitude=-86.6286,
element='TMIN', start=1897, end=2024),
Inventory(station='USC00478937', latitude=42.9986, longitude=-88.2525,
element='TMAX', start=1894, end=2024),
Inventory(station='USC00478937', latitude=42.9986, longitude=-88.2525,
element='TMIN', start=1894, end=2024)]Selecting a station and getting the station metadata
As you look at the top 20 entries in your newly sorted list, it seems that the first station, USC00110338, is a good fit. It’s got both TMIN and TMAX and one of the longer series, starting in 1893 and running up through 2017, for more than 120 years’ worth of data. So save that station into your station variable and quickly parse the station data you’ve already grabbed to pick up a little more information about the station.
Back in the readme file, you find the following information about the station data:
IV. FORMAT OF "ghcnd-stations.txt"
------------------------------
Variable Columns Type
------------------------------
ID 1-11 Character
LATITUDE 13-20 Real
LONGITUDE 22-30 Real
ELEVATION 32-37 Real
STATE 39-40 Character
NAME 42-71 Character
GSN FLAG 73-75 Character
HCN/CRN FLAG 77-79 Character
WMO ID 81-85 Character
------------------------------
These variables have the following definitions:
ID is the station identification code. Note that the first two
characters denote the FIPS country code, the third character
is a network code that identifies the station numbering system
used, and the remaining eight characters contain the actual
station ID.
See "ghcnd-countries.txt" for a complete list of country codes.
See "ghcnd-states.txt" for a list of state/province/territory codes.
The network code has the following five values:
0 = unspecified (station identified by up to eight
alphanumeric characters)
1 = Community Collaborative Rain, Hail,and Snow (CoCoRaHS)
based identification number. To ensure consistency with
with GHCN Daily, all numbers in the original CoCoRaHS IDs
have been left-filled to make them all four digits long.
In addition, the characters "-" and "_" have been removed
to ensure that the IDs do not exceed 11 characters when
preceded by "US1". For example, the CoCoRaHS ID
"AZ-MR-156" becomes "US1AZMR0156" in GHCN-Daily
C = U.S. Cooperative Network identification number (last six
characters of the GHCN-Daily ID)
E = Identification number used in the ECA&D non-blended
dataset
M = World Meteorological Organization ID (last five
characters of the GHCN-Daily ID)
N = Identification number used in data supplied by a
National Meteorological or Hydrological Center
R = U.S. Interagency Remote Automatic Weather Station (RAWS)
identifier
S = U.S. Natural Resources Conservation Service SNOwpack
TELemtry (SNOTEL) station identifier
W = WBAN identification number (last five characters of the
GHCN-Daily ID)
LATITUDE is latitude of the station (in decimal degrees).
LONGITUDE is the longitude of the station (in decimal degrees).
ELEVATION is the elevation of the station (in meters, missing = -999.9).
STATE is the U.S. postal code for the state (for U.S. stations only).
NAME is the name of the station.
GSN FLAG is a flag that indicates whether the station is part of the GCOS
Surface Network (GSN). The flag is assigned by cross-referencing
the number in the WMOID field with the official list of GSN
stations. There are two possible values:
Blank = non-GSN station or WMO Station number not available
GSN = GSN station
HCN/ is a flag that indicates whether the station is part of the U.S.
CRN FLAG Historical Climatology Network (HCN). There are three possible
values:
Blank = Not a member of the U.S. Historical Climatology
or U.S. Climate Reference Networks
HCN = U.S. Historical Climatology Network station
CRN = U.S. Climate Reference Network or U.S. Regional Climate
Network Station
WMO ID is the World Meteorological Organization (WMO) number for the
station. If the station has no WMO number (or one has not yet
been matched to this station), then the field is blank.Although you might care more about the metadata fields for more serious research, right now you want to match the start and end year from the inventory records to the rest of the station metadata in the stations file.
You have several ways to sift through the stations file to find the one station that matches the station ID you selected. You could create a for loop to go through each line and break out when you find it; you could split the data into lines and then sort and use a binary search; and so on. Depending on the nature and amount of data you have, one approach or another might be appropriate. In this case, because you have the data loaded already, and it’s not too large, use a list comprehension to return a list with its single element being the station you’re looking for:
station_id = 'USC00110338'
# parse stations
Station = namedtuple("Station", ['station_id', 'latitude', 'longitude',
'elevation', 'state', 'name', 'start', 'end'])
stations = [(x[0:11], float(x[12:20]), float(x[21:30]), float(x[31:37]),
x[38:40].strip(), x[41:71].strip())
for x in stations_txt.split("\n") if x.startswith(station_id)]
station = Station(*stations[0] + (inventory_temps[0].start,
inventory_temps[0].end))
print(station)Station(station_id='USC00110338', latitude=41.7803, longitude=-88.3092,
elevation=205.7, state='IL', name='AURORA WATER', start=1915, end=2024)At this point, you’ve identified that you want weather data from the station at Aurora, Illinois, which is the nearest station to downtown Chicago with more than a century’s worth of temperature data.
Fetching and parsing the actual weather data
With the station identified, the next step is fetching the actual weather data for that station and parsing it. The process is quite similar to what you did in the preceding section.
Fetching the data
First, fetch the data file and save it, in case you need to go back to it:
# fetch daily records for selected station
r = requests.get('https://www1.ncdc.noaa.gov/pub/data/ghcn/daily/all/{}.dly'
➥.format(station.station_id))
weather = r.text
# save into a text file, so we won't need to fetch again
with open('weather_{}.txt'.format(station), "w") as weather_file:
weather_file.write(weather)# read from saved daily file if needed (only used if we want to start the
➥process over without downloadng the file)
with open('weather_{}.txt'.format(station)) as weather_file:
weather = weather_file.read()
print(weather[:540])
USC00110338189301TMAX -11 6 -44 6 -139 6 -83 6 -100 6 -83 6 -72
➥6 -83 6 -33 6 -178 6 -150 6 -128 6 -172 6 -200 6 -189 6 -150
➥6 -106 6 -61 6 -94 6 -33 6 -33 6 -33 6 -33 6 6 6 -33
➥6 -78 6 -33 6 44 6 -89 I6 -22 6 6 6
USC00110338189301TMIN -50 6 -139 6 -250 6 -144 6 -178 6 -228 6 -144
➥6 -222 6 -178 6 -250 6 -200 6 -206 6 -267 6 -272 6 -294 6 -294
➥6 -311 6 -200 6 -233 6 -178 6 -156 6 -89 6 -200 6 -194 6 -194
➥6 -178 6 -200 6 -33 I6 -156 6 -139 6 -167 6Parsing the weather data
Again, now that you have the data, you can see it’s quite a bit more complex than the station and inventory data. Clearly, it’s time to head back to the readme.txt file and section III, which is the description of a weather data file. You have a lot of options, so filter them down to the ones that concern you; leave out the other element types as well as the whole system of flags specifying the source, quality, and type of the values:
III. FORMAT OF DATA FILES (".dly" FILES)
Each ".dly" file contains data for one station. The name of the file
corresponds to a station's identification code. For example, "USC00026481.dly"
contains the data for the station with the identification code USC00026481).
Each record in a file contains one month of daily data. The variables on each
line include the following:
------------------------------
Variable Columns Type
------------------------------
ID 1-11 Character
YEAR 12-15 Integer
MONTH 16-17 Integer
ELEMENT 18-21 Character
VALUE1 22-26 Integer
MFLAG1 27-27 Character
QFLAG1 28-28 Character
SFLAG1 29-29 Character
VALUE2 30-34 Integer
MFLAG2 35-35 Character
QFLAG2 36-36 Character
SFLAG2 37-37 Character
. . .
. . .
. . .
VALUE31 262-266 Integer
MFLAG31 267-267 Character
QFLAG31 268-268 Character
SFLAG31 269-269 Character
------------------------------
These variables have the following definitions:
ID is the station identification code. Please see "ghcnd-stations.txt"
for a complete list of stations and their metadata.
YEAR is the year of the record.
MONTH is the month of the record.
ELEMENT is the element type. There are five core elements as well as a number
of addition elements.
The five core elements are:
PRCP = Precipitation (tenths of mm)
SNOW = Snowfall (mm)
SNWD = Snow depth (mm)
TMAX = Maximum temperature (tenths of degrees C)
TMIN = Minimum temperature (tenths of degrees C)
...
VALUE1 is the value on the first day of the month (missing = -9999).
MFLAG1 is the measurement flag for the first day of the month.
QFLAG1 is the quality flag for the first day of the month.
SFLAG1 is the source flag for the first day of the month.
VALUE2 is the value on the second day of the month
MFLAG2 is the measurement flag for the second day of the month.
QFLAG2 is the quality flag for the second day of the month.
SFLAG2 is the source flag for the second day of the month.
... and so on through the 31st day of the month. Note: If the month has less
than 31 days, then the remaining variables are set to missing (e.g., for April,
VALUE31 = -9999, MFLAG31 = blank, QFLAG31 = blank, SFLAG31 = blank).The key points you care about right now are that the station ID is the 11 characters of a row; the year is the next 4, the month the next 2, and the element the next 4 after that. After that, there are 31 slots for daily data, with each slot consisting of 5 characters for the temperature, expressed in tenths of a degree Celsius, and 3 characters of flags. As I mentioned earlier, you can disregard the flags for this exercise. You can also see that missing values for the temperatures are coded with –9999 if that day isn’t in the month, so for a typical February, for example, the 29th, 30th, and 31st values would be –9999.
As you process your data in this exercise, you’re looking to get overall trends, so you don’t need to worry much about individual days. Instead, find average values for the month. You can save the maximum, minimum, and mean values for the entire month and use those.
This means that to process each line of weather data, you need to
- 1 Split the line into its separate fields and ignore or discard the flags for each daily value.
- 2 Remove the values with –9999, and convert the year and month into ints and the temperature values into floats, keeping in mind that the temperature readings are in tenths of degrees centigrade.
- 3 Calculate the average value, and pick out the high and low values.
To accomplish all these tasks, you can take a couple of approaches. You could do several passes over the data, splitting into fields, discarding the placeholders, converting strings to numbers, and, finally, calculating the summary values. Or you can write a function that performs all of these operations on a single line and do everything in one pass. Both approaches can be valid. In this case, take the latter approach and create a parse_line function to perform all of your data transformations:
def parse_line(line):
""" parses line of weather data
removes values of -9999 (missing value)
"""
# return None if line is empty
if not line:
return None
# split out first 4 fields and string containing temperature values
record, temperature_string = (line[:11], int(line[11:15]),
int(line[15:17]), line[17:21]), line[21:]
# raise exception if the temperature string is too short
if len(temperature_string) < 248:
raise ValueError("String not long enough - {} {}".format(temperature_
string, str(line)))
# use a list comprehension on the temperature_string to extract and
➥convert the
values = [float(temperature_string[i:i + 5])/10 for i in range(0, 248, 8)
if not temperature_string[i:i + 5].startswith("-9999")]
# get the number of values, the max and min, and calculate average count = len(values)
tmax = round(max(values), 1)
tmin = round(min(values), 1)
mean = round(sum(values)/count, 1)
# add the temperature summary values to the record fields extracted
➥earlier and return
return record + (tmax, tmin, mean, count)If you test this function with the first line of your raw weather data, you get the following result:
parse_line(weather[:270])
('USC00110338', 1893, 1, 'TMAX', 4.4, -20.0, -7.8, 31)So it looks like you have a function that will work to parse your data. If that function works, you can parse the weather data and either store it or continue with your processing:
# process all weather data
# list comprehension, will not parse empty lines
weather_data = [parse_line(x) for x in weather.split("\n") if x]
len(weather_data)
weather_data[:10]
[('USC00110338', 1893, 1, 'TMAX', 4.4, -20.0, -7.8, 31),
('USC00110338', 1893, 1, 'TMIN', -3.3, -31.1, -19.2, 31),
('USC00110338', 1893, 1, 'PRCP', 8.9, 0.0, 1.1, 31),
('USC00110338', 1893, 1, 'SNOW', 10.2, 0.0, 1.0, 31),
('USC00110338', 1893, 1, 'WT16', 0.1, 0.1, 0.1, 2),
('USC00110338', 1893, 1, 'WT18', 0.1, 0.1, 0.1, 11),
('USC00110338', 1893, 2, 'TMAX', 5.6, -17.2, -0.9, 27),
('USC00110338', 1893, 2, 'TMIN', 0.6, -26.1, -11.7, 27),
('USC00110338', 1893, 2, 'PRCP', 15.0, 0.0, 2.0, 28),
('USC00110338', 1893, 2, 'SNOW', 12.7, 0.0, 0.6, 28)]Now you have all the weather records, not just the temperature records, parsed and in your list.
Saving the weather data in a database (optional)
At this point, you can save all of the weather records (and the station records and inventory records as well if you want) in a database. Doing so lets you come back in later sessions and use the same data without having to go to the hassle of fetching and parsing the data again.
As an example, the following code is how you could save the weather data in a sqlite3 database:
import sqlite3
conn = sqlite3.connect("weather_data.db")
cursor = conn.cursor()
# create weather table
create_weather = """CREATE TABLE "weather" (
"id" text NOT NULL,
"year" integer NOT NULL,
"month" integer NOT NULL,
"element" text NOT NULL,
"max" real,
"min" real,
"mean" real,
"count" integer)"""
cursor.execute(create_weather)
conn.commit()
# store parsed weather data in database
for record in weather_data:
cursor.execute("""insert into weather (id, year, month, element, max,
➥min, mean, count) values (?,?,?,?,?,?,?,?) """,
record)
conn.commit()When you have the data stored, you could retrieve it from the database with code like the following, which fetches only the TMAX records:
cursor.execute("""select * from weather where element='TMAX' order by year
➥ month""")
tmax_data = cursor.fetchall()
tmax_data[:5]
[('USC00110338', 1893, 1, 'TMAX', 4.4, -20.0, -7.8, 31),
('USC00110338', 1893, 2, 'TMAX', 5.6, -17.2, -0.9, 27),
('USC00110338', 1893, 3, 'TMAX', 20.6, -7.2, 5.6, 30),
('USC00110338', 1893, 4, 'TMAX', 28.9, 3.3, 13.5, 30),
('USC00110338', 1893, 5, 'TMAX', 30.6, 7.2, 19.2, 31)]Selecting and graphing data
Because you’re concerned only with temperature, you need to select just the temperature records. You can do that quickly enough by using a couple of list comprehensions to pick out a list for TMAX and one for TMIN. Or you could use the features of pandas, which you’ll be using for graphing the data, to filter out the records you don’t want. Because you’re more concerned with pure Python than with pandas, take the first approach:
tmax_data = [x for x in weather_data if x[3] == ‘TMAX’] tmin_data = [x for x in weather_data if x[3] == ‘TMIN’]
tmin_data[:5]
[('USC00110338', 1893, 1, 'TMIN', -3.3, -31.1, -19.2, 31),
('USC00110338', 1893, 2, 'TMIN', 0.6, -26.1, -11.7, 27),
('USC00110338', 1893, 3, 'TMIN', 3.3, -13.3, -4.6, 31),
('USC00110338', 1893, 4, 'TMIN', 12.2, -5.6, 2.2, 30),
('USC00110338', 1893, 5, 'TMIN', 14.4, -0.6, 5.7, 31)]Using pandas to graph your data
At this point, you have your data cleaned and ready to graph. To make the graphing easier, you can use pandas and matplotlib, as described in chapter 24. If you are using Colaboratory, both are already installed and available. If you are using another environment, you could install them with the following commands at the command line:
pip install pandas matplotlibTo use pandas, we can import it as usual, but to use matplotlib in Colaboratory, we can just use the Jupyter “magic” command to make it available:
import pandas as pd %matplotlib inline Magic command to activate matplotib
Then you can load pandas and create data frames for your TMAX and TMIN data:
tmax_df = pd.DataFrame(tmax_data, columns=['Station', 'Year', 'Month',
'Element', 'Max', 'Min', 'Mean', 'Days'])
tmin_df = pd.DataFrame(tmin_data, columns=['Station', 'Year', 'Month',
'Element', 'Max', 'Min', 'Mean', 'Days'])You could plot the monthly values, but 125 years times 12 months of data is 1,500 data points, and the cycle of seasons also makes picking out patterns difficult.
Instead, it probably makes more sense to average the high, low, and mean monthly values into yearly values and plot those values. You could do this in Python, but because you already have your data loaded in a pandas data frame, you can use that to group by year and get the mean values:
# select Year, Min, Max, Mean columns, group by year, average and line plot
tmin_df[['Year','Min', 'Mean', 'Max']].groupby('Year').mean().plot(
kind='line', figsize=(16, 8))This result has a fair amount of variation, but it does seem to indicate that the minimum temperature has been on the rise for the past 20 years.
Note that if you wanted to get the same graph without using Jupyter notebook and matplotlib, you could use still use pandas, but you’d write to a CSV or Microsoft Excel file, using the data frame’s to_csv or to_excel method. Then you could load the resulting file into a spreadsheet and graph from there.
appendix A guide to Python’s documentation
The best and most current reference for Python is the documentation that comes with Python itself. With that in mind, it’s more useful to explore the ways you can access that documentation than to print pages of edited documentation.
The standard bundle of documentation has several sections, including instructions on documenting, distributing, installing, and extending Python on various platforms, and is the logical starting point when you’re looking for answers to questions about Python. The two main areas of the Python documentation that are likely to be the most useful are the “Library Reference” and the “Language Reference.” The “Library Reference” is absolutely essential because it has explanations of both the built-in data types and every module included with Python. The “Language Reference” is the explanation of how the core of Python works, and it contains the official word on the core of the language, explaining the workings of data types, statements, and so on. The “What’s New” section is also worth reading, particularly when a new version of Python is released, because it summarizes all of the changes in the new version.
A.1 Accessing Python documentation on the web
For many people, the most convenient way to access the Python documentation is to go to docs.python.org and browse the documentation collection there. Although doing so requires a connection to the web, it has the advantage that the docs for the most current version are available, as well as the docs for all previous versions. Given that, for many projects, it’s often useful to search the web for other documentation and information, having a browser tab permanently open and pointing to the online Python documentation is an easy way to have a Python reference at your fingertips.
If you want the Python documentation saved locally on a computer, tablet, or e-book reader, you can also download the complete documentation from docs.python.org in PDF, HTML plain text. Texinfo, and EPUB formats.
A.2 Best practices: How to become a Pythonista
Every programming language develops its own traditions and culture, and Python is a strong example. Most experienced Python programmers (Pythonistas, as they’re sometimes called) care a great deal about writing Python in a way that matches the style and best practices of Python. This type of code is commonly called Pythonic code and is valued highly, as opposed to Python code that looks like Java, C, or JavaScript.
The challenge that coders new to Python face is how to learn to write Pythonic code. Although getting a feel for the language and its style takes a little time and effort, the rest of this appendix gives you some suggestions on how to start.
A.2.1 Ten tips for becoming a Pythonista
The tips in this section are ones that I share with intermediate Python classes and are my suggestions for leveling up your Python skills. I’m not saying that everyone absolutely agrees with me, but from what I’ve seen over the years, these tips will put you soundly on the path to being a true Pythonista:
- Consider The Zen of Python. “The Zen of Python,” or Python Enhancement Proposal (PEP) 20, sums up the design philosophy underlying Python as a language and is commonly invoked in discussions of what makes scripts more Pythonic. In particular, “Beautiful is better than ugly” and “Simple is better than complex” should guide your coding. I’ve included “The Zen of Python” at the end of this appendix; you can always find it by typing import this at a Python shell prompt.
- Follow PEP 8. PEP 8 is the official Python style guide, which is also included later in this appendix. PEP 8 offers good advice on everything from code formatting and variable naming to the use of the language. If you want to write Pythonic code, become familiar with PEP 8.
- Be familiar with the docs. Python has a rich, well-maintained collection of documentation, and you should refer to it often. The most useful documents probably are the standard library documentation, but the tutorials and how-to files are also rich veins of information on using the language effectively.
- Write as little code as you can as much as you can. Although this advice might apply to many languages, it fits Python particularly well. What I mean is that you should strive to make your programs as short and as simple as possible (but no shorter and no simpler) and that you should practice that style of coding as much as you can.
- Read as much code as you can. From the beginning, the Python community has been aware that reading code is more important than writing code. Read as much Python code as you can, and if possible, discuss the code that you read with others.
- Use the built-in data structures over all else. You should turn first to Python’s built-in structures before writing your own classes to hold data. Python’s various data types can be combined with nearly unlimited flexibility and have the advantage of years of debugging and optimization. Take advantage of them.
- Dwell on generators and comprehensions. Coders who are new to Python almost always fail to appreciate how much list and dictionary comprehensions and generator expressions are a part of Pythonic coding. Look at examples in the Python code that you read, and practice them. You won’t be a Pythonista until you can write a list comprehension almost without thinking.
- Use the standard library. When the built-ins fail you, look next to the standard library. The elements in the standard library are the famed “batteries included” of Python. They’ve stood the test of time and have been optimized and documented better than almost any other Python code. Use them if you can.
- Write as few classes as you can. Write your own classes only if you must. Experienced Pythonistas tend to be very economical with classes, knowing that designing good classes isn’t trivial and that any classes they create are also classes that they have to test and debug.
- Be wary of frameworks. Frameworks can be attractive, particularly to coders new to the language, because they offer so many powerful shortcuts. You should use frameworks when they’re helpful, of course, but be aware of their downsides. You may spend more time learning the quirks of an un-Pythonic framework than learning Python itself, or you may find yourself adapting what you do to the framework rather than the other way around.
A.3 PEP 8: Style guide for Python code
This section contains a slightly edited excerpt from PEP 8. Written by Guido van Rossum and Barry Warsaw, PEP 8 is the closest thing Python has to a style manual. Some more-specific sections have been omitted, but the main points are covered. You should make your code conform to PEP 8 as much as possible; your Python style will be the better for it.
You can access the full text of PEP 8 and all of the other PEPs put forth in the history of Python by going to the documentation section of peps.python.org and looking at the PEP index. The PEPs are excellent sources for the history and lore of Python as well as explanations of current concerns and future plans.
A.3.1 Introduction
This document gives coding conventions for the Python code comprising the standard library in the main Python distribution. Please see the companion informational PEP describing style guidelines for the C code in the C implementation of Python.1 This document was adapted from Guido’s original “Python Style Guide” essay, with some additions from Barry’s style guide.2 Where there’s conflict, Guido’s style rules for the purposes of this PEP. This PEP may still be incomplete (in fact, it may never be finished <wink>).
A foolish consistency is the hobgoblin of little minds
One of Guido’s key insights is that code is read much more often than it’s written. The guidelines provided here are intended to improve the readability of code and make it consistent across the wide spectrum of Python code. As PEP 20 says, “Readability counts.”3
A style guide is about consistency. Consistency with this style guide is important; consistency within a project is more important; consistency within one module or function is even more important.
But the most important thing is to know when to be inconsistent—sometimes the style guide just doesn’t apply. When in doubt, use your best judgment. Look at other examples and decide what looks best. And don’t hesitate to ask!
The following are two good reasons to break a particular rule:
- When applying the rule would make the code less readable, even for someone who is used to reading code that follows the rules
- To be consistent with surrounding code that also breaks it (maybe for historic reasons), although this is also an opportunity to clean up someone else’s mess (in true XP style)
A.3.2 Code layout
Indentation
Use four spaces per indentation level.
For really old code that you don’t want to mess up, you can continue to use eightspace tabs.
Tabs or spaces?
Never mix tabs and spaces.
The most popular way of indenting Python is with spaces only. The second most popular way is with tabs only. Code indented with a mixture of tabs and spaces should be converted to using spaces exclusively. When you invoke the Python command-line interpreter with the -t option, it puts forth warnings about code that illegally mixes tabs and spaces. When you use -tt, these warnings become errors. These options are highly recommended!
1 PEP 7, “Style Guide for C Code,” van Rossum, https://www.python.org/dev/peps/pep-0007/.
2 Barry Warsaw’s “GNU Mailman Coding Style Guide,” http://barry.warsaw.us/software/STYLEGUIDE.txt. The URL is empty although it is presented in the PEP 8 style guide.
3 PEP 20, “The Zen of Python,” www.python.org/dev/peps/pep-0020/.
For new projects, spaces only are strongly recommended over tabs. Most editors have features that make this easy to do.
Maximum line length
Limit all lines to a maximum of 79 characters.
Many devices are still around that are limited to 80-character lines; plus, limiting windows to 80 characters makes it possible to have several windows side by side. The default wrapping on such devices disrupts the visual structure of the code, making it more difficult to understand. Therefore, please limit all lines to a maximum of 79 characters. For flowing long blocks of text (docstrings or comments), limiting the length to 72 characters is recommended.
The preferred way of wrapping long lines is by using Python’s implied line continuation inside parentheses, brackets, and braces. If necessary, you can add an extra pair of parentheses around an expression, but sometimes using a backslash looks better. Make sure to indent the continued line appropriately. The preferred place to break around a binary operator is after the operator, not before it. The following are some examples:
class Rectangle(Blob):
def __init__(self, width, height,
color='black', emphasis=None, highlight=0):
if width == 0 and height == 0 and \
color == 'red' and emphasis == 'strong' or \
highlight > 100:
raise ValueError("sorry, you lose")
if width == 0 and height == 0 and (color == 'red' or \
emphasis is None):
raise ValueError("I don't think so -- values are %s, %s" % \
(width, height))
Blob.__init__(self, width, height, \
color, emphasis, highlight)Blank lines
Separate top-level function and class definitions with two blank lines. Method definitions inside a class are separated by a single blank line. Extra blank lines may be used (sparingly) to separate groups of related functions. Blank lines may be omitted between a bunch of related one-liners (for example, a set of dummy implementations). Use blank lines in functions, sparingly, to indicate logical sections.
Python accepts the Ctrl-L (^L) form feed character as whitespace. Many tools treat these characters as page separators, so you may use them to separate pages of related sections of your file.
Imports
Imports should usually be on separate lines—for example:
import os
import sysDon’t put them together like this:
import sys, osIt’s okay to say this, though:
from subprocess import Popen, PIPEImports are always put at the top of the file, just after any module comments and docstrings and before module globals and constants.
Imports should be grouped in the following order:
- 1 Standard library imports
- 2 Related third-party imports
- 3 Local application/library–specific imports
Put a blank line between each group of imports. Put any relevant __all__ specifications after the imports.
Relative imports for intrapackage imports are highly discouraged. Always use the absolute package path for all imports. Even now that PEP 3284 is fully implemented in Python 2.5, its style of explicit relative imports is actively discouraged; absolute imports are more portable and usually more readable.
When importing a class from a class-containing module, it’s usually okay to spell them as follows:
from myclass import MyClass
from foo.bar.yourclass import YourClassIf this spelling causes local name clashes, then spell them as follows:
import myclass
import foo.bar.yourclass
and use myclass.MyClass and foo.bar.yourclass.YourClass.Whitespace in expressions and statements
Here are some of my pet peeves. Avoid extraneous whitespace in the following situations:
Immediately inside parentheses, brackets, or braces
Yes:spam(ham[1], {eggs: 2})No:
spam( ham[ 1 ], { eggs: 2 } )
4 PEP 328, “Imports: Multi-Line and Absolute/Relative,” www.python.org/dev/peps/pep-0328/.
Immediately before a comma, semicolon, or colon
Yes:if x == 4: print x, y; x, y = y, xNo:
if x == 4 : print x , y ; x , y = y , xImmediately before the open parenthesis that starts the argument list of a function call
Yes:dict['key'] = list[index]No:
dict ['key'] = list [index]More than one space around an assignment (or other) operator to align it with another
Yes:x = 1 y = 2 long_variable = 3No:
x = 1 y = 2 long_variable = 3
Other recommendations
Always surround these binary operators with a single space on either side: assignment (=), augmented assignment (+=, -=, and so on), comparisons (==, <, >, !=, <>, <=, >=, in, not in, is, is not), and Booleans (and, or, not).
Use spaces around arithmetic operators.
Yes:
i = i + 1
submitted += 1
x = x * 2 – 1
hypot2 = x * x + y * y
c = (a + b) * (a - b)No:
i=i+1
submitted +=1
x = x*2 – 1
hypot2 = x*x + y*y
c = (a+b) * (a-b)Don’t use spaces around the = sign when used to indicate a keyword argument or a default parameter value.
Yes:
def complex(real, imag=0.0):
return magic(r=real, i=imag)No:
def complex(real, imag = 0.0):
return magic(r = real, i = imag)Compound statements (multiple statements on the same line) are generally discouraged.
Yes:
if foo == 'blah':
do_blah_thing()
do_one()
do_two()
do_three()Rather not:
if foo == 'blah': do_blah_thing()
do_one(); do_two(); do_three()While sometimes it’s okay to put an if/for/while with a small body on the same line, never do this for multiclause statements. Also avoid folding such long lines!
Rather not:
if foo == 'blah': do_blah_thing()
for x in lst: total += x
while t < 10: t = delay()No:
if foo == 'blah': do_blah_thing()
else: do_non_blah_thing()
try: something()
finally: cleanup()
do_one(); do_two(); do_three(long, argument,
list, like, this)
if foo == 'blah': one(); two(); three()A.4 Comments
Comments that contradict the code are worse than no comments. Always make a priority of keeping the comments up to date when the code changes!
Comments should be complete sentences. If a comment is a phrase or sentence, its first word should be capitalized, unless it’s an identifier that begins with a lowercase letter (never alter the case of identifiers!).
If a comment is short, the period at the end can be omitted. Block comments generally consist of one or more paragraphs built out of complete sentences, and each sentence should end in a period. Use two spaces after a sentence-ending period. When writing English, Strunk & White applies.
If you are a Python coder from a non-English-speaking country, you should write your comments in English, unless you are 120% sure that the code will never be read by people who don’t speak your language.
Block comments
Block comments generally apply to some (or all) code that follows them and are indented to the same level as that code. Each line of a block comment starts with a # and a single space (unless it is indented text inside the comment).
Paragraphs inside a block comment are separated by a line containing a single #.
Inline comments
Use inline comments sparingly. An inline comment is a comment on the same line as a statement. Inline comments should be separated by at least two spaces from the statement. They should start with a # and a single space.
Inline comments are unnecessary and in fact distracting if they state the obvious. Don’t do this:
x = x + 1 # Increment x
But sometimes, this is useful:
x = x + 1 # Compensate for border
Documentation strings
Conventions for writing good documentation strings (docstrings) are immortalized in PEP 257.5
Write docstrings for all public modules, functions, classes, and methods. Docstrings are not necessary for nonpublic methods, but you should have a comment that describes what the method does. This comment should appear after the def line.
PEP 257 describes good docstring conventions. Note that, most importantly, the ““” that ends a multiline docstring should be on a line by itself and preferably preceded by a blank line—for example:
"""Return a foobang
Optional plotz says to frobnicate the bizbaz first.
"""For one-liner docstrings, it’s okay to keep the closing ““” on the same line.
Version bookkeeping
If you have to have Subversion, CVS, or RCS crud in your source file, do it as follows:
__version__ = "$Revision: 68852 $" # $Source$These lines should be included after the module’s docstring, before any other code, separated by a blank line above and below.
A.4.1 Naming conventions
The naming conventions of Python’s library are a bit of a mess, so we’ll never get this completely consistent. Nevertheless, the following are the currently recommended naming standards. New modules and packages (including third-party frameworks) should be written to these standards, but where an existing library has a different style, internal consistency is preferred.
Descriptive: Naming styles
There are many different naming styles. It helps to be able to recognize what naming style is being used, independent of what it’s used for.
The following naming styles are commonly distinguished:
- b (single lowercase letter)
- B (single uppercase letter)
- lowercase
- lower_case_with_underscores
- UPPERCASE
- UPPER_CASE_WITH_UNDERSCORES
- CapitalizedWords (or CapWords, or CamelCase—so named because of the bumpy look of its letters). This is also sometimes known as StudlyCaps.
- Note: When using abbreviations in CapWords, capitalize all the letters of the abbreviation. Thus HTTPServerError is better than HttpServerError.
- mixedCase (differs from CapitalizedWords by initial lowercase character!)
- Capitalized_Words_With_Underscores (ugly!)
5 PEP 257, “Docstring Conventions,” Goodger and van Rossum, www.python.org/dev/peps/pep-0257/.
There’s also the style of using a short unique prefix to group related names together. This is seldom used in Python, but I mention it for completeness. For example, the os.stat() function returns a tuple whose items traditionally have names like st_mode, st_size, st_mtime, and so on. (This is done to emphasize the correspondence with the fields of the POSIX system call struct, which helps programmers familiar with that.)
The X11 library uses a leading X for all its public functions. In Python, this style is generally deemed unnecessary because attribute and method names are prefixed with an object, and function names are prefixed with a module name.
In addition, the following special forms using leading or trailing underscores are recognized (these can generally be combined with any case convention):
_single_leading_underscore
Weak “internal use” indicator. For example, from M import * does not import objects whose name starts with an underscore.single_trailing_underscore_
Used by convention to avoid conflicts with Python keyword—for example:tkinter.Toplevel(master, class\_='ClassName').__double_leading_underscore
When naming a class attribute, it invokes name mangling (inside class FooBar, __boo becomes _FooBar__boo; see later discussion).__double_leading_and_trailing_underscore__ “Magic” objects or attributes that live in user-controlled namespaces. For example, __init__, __import__ or __file__. Never invent such names; use them only as documented.
Prescriptive: Naming conventions
Names to avoid
Never use the characters l (lowercase letter el), O (uppercase letter oh), or I (uppercase letter eye) as single-character variable names.
In some fonts, these characters are indistinguishable from the numerals 1 (one) and 0 (zero). When tempted to use l, use L instead.
Package and module names
Modules should have short, all-lowercase names. Underscores can be used in a module name if it improves readability. Python packages should also have short, all-lowercase names, although the use of underscores is discouraged.
Since module names are mapped to filenames, and some filesystems are case insensitive and truncate long names, it’s important that module names be fairly short—this won’t be a problem on UNIX, but it may be a problem when the code is transported to older Mac or Windows versions or DOS.
When an extension module written in C or C++ has an accompanying Python module that provides a higher-level (for example, more object-oriented) interface, the C/C++ module has a leading underscore (for example, _socket).
Class names
Almost without exception, class names use the CapWords convention. Classes for internal use have a leading underscore in addition.
Exception names
Because exceptions should be classes, the class-naming convention applies here. However, you should use the suffix Error on your exception names (if the exception actually is an error).
Global variable names
(Let’s hope that these variables are meant for use inside one module only.) The conventions are about the same as those for functions.
Modules that are designed for use via from M import * should use the __all__ mechanism to prevent exporting globals or use the older convention of prefixing such globals with an underscore (which you might want to do to indicate these globals are module nonpublic).
Function names
Function names should be lowercase, with words separated by underscores as necessary to improve readability.
mixedCase is allowed only in contexts where that’s already the prevailing style (for example, threading.py), to retain backward compatibility.
Function and method arguments
Always use self for the first argument to instance methods.
Always use cls for the first argument to class methods.
If a function argument’s name clashes with a reserved keyword, it’s generally better to append a single trailing underscore than to use an abbreviation or spelling corruption. Thus, print_ is better than prnt. (Perhaps better is to avoid such clashes by using a synonym.)
Method names and instance variables
Use the function-naming rules: lowercase with words separated by underscores as necessary to improve readability.
Use one leading underscore only for nonpublic methods and instance variables.
To avoid name clashes with subclasses, use two leading underscores to invoke Python’s name-mangling rules.
Python mangles these names with the class name: if class Foo has an attribute named __a, it cannot be accessed by Foo.__a. (An insistent user could still gain access by calling Foo._Foo__a.) Generally, double leading underscores should be used only to avoid name conflicts with attributes in classes designed to be subclassed.
Note: There is some controversy about the use of __names.
Constants
Constants are usually declared on a module level and written in all capital letters with underscores separating words. Examples include MAX_OVERFLOW and TOTAL.
Designing for inheritance
Always decide whether a class’s methods and instance variables (collectively called attributes) should be public or nonpublic. If in doubt, choose nonpublic; it’s easier to make it public later than to make a public attribute nonpublic.
Public attributes are those that you expect unrelated clients of your class to use, with your commitment to avoid backward-incompatible changes. Nonpublic attributes are those that are not intended to be used by third parties; you make no guarantees that nonpublic attributes won’t change or even be removed.
We don’t use the term private here, since no attribute is really private in Python (without a generally unnecessary amount of work).
Another category of attributes includes those that are part of the subclass API (often called protected in other languages). Some classes are designed to be inherited from, either to extend or modify aspects of the class’s behavior. When designing such a class, take care to make explicit decisions about which attributes are public, which are part of the subclass API, and which are truly only to be used by your base class.
With this in mind, here are the Pythonic guidelines:
Public attributes should have no leading underscores.
If your public attribute name collides with a reserved keyword, append a single trailing underscore to your attribute name. This is preferable to an abbreviation or corrupted spelling. (However, notwithstanding this rule, cls is the preferred spelling for any variable or argument that’s known to be a class, especially the first argument to a class method.)
Note 1: See the previous argument name recommendation for class methods.For simple public data attributes, it’s best to expose just the attribute name, without complicated accessor/mutator methods. Keep in mind that Python provides an easy path to future enhancement should you find that a simple data attribute needs to grow functional behavior. In that case, use properties to hide functional implementation behind simple data attribute access syntax.
Note 1: Properties work only on new-style classes.
Note 2: Try to keep the functional behavior side-effect free, although side effects such as caching are generally fine.
Note 3: Avoid using properties for computationally expensive operations; the attribute notation makes the caller believe that access is (relatively) cheap.If your class is intended to be subclassed, and you have attributes that you don’t want subclasses to use, consider naming them with double leading underscores and no trailing underscores. This invokes Python’s name-mangling algorithm, where the name of the class is mangled into the attribute name. This helps avoid attribute name collisions should subclasses inadvertently contain attributes with the same name.
Note 1: Only the simple class name is used in the mangled name, so if a subclass chooses both the same class name and attribute name, you can still get name collisions.
Note 2: Name mangling can make certain uses, such as debugging and __getattr__(), less convenient. However, the name-mangling algorithm is well documented and easy to perform manually.
Note 3: Not everyone likes name mangling. Try to balance the need to avoid accidental name clashes with potential use by advanced callers.
A.4.2 Programming recommendations
You should write code in a way that does not disadvantage other implementations of Python (PyPy, Jython, IronPython, Pyrex, Psyco, and such).
For example, don’t rely on CPython’s efficient implementation of in-place string concatenation for statements in the form a+=b or a=a+b. Those statements run more slowly in Jython. In performance-sensitive parts of the library, you should use the ’’.join() form instead. This will ensure that concatenation occurs in linear time across various implementations.
Comparisons to singletons like None should always be done with is or is not—never the equality operators.
Also, beware of writing if x when you really mean if x is not None—for example, when testing whether a variable or argument that defaults to None was set to some other value. The other value might have a type (such as a container) that could be false in a Boolean context!
Use class-based exceptions. String exceptions in new code are forbidden, because this language feature was removed in Python 2.6.
Modules or packages should define their own domain-specific base exception class, which should be subclassed from the built-in Exception class. Always include a class docstring, for example:
class MessageError(Exception):
"""Base class for errors in the email package."""Class-naming conventions apply here, although you should add the suffix Error to your exception classes if the exception is an error. Nonerror exceptions need no special suffix.
When raising an exception, use raise ValueError(‘message’) instead of the older form raise ValueError, ‘message’.
The parenthetical form is preferred because, when the exception arguments are long or include string formatting, you don’t need to use line continuation characters, thanks to the containing parentheses. The older form was removed in Python 3.
When catching exceptions, mention specific exceptions whenever possible instead of using a bare except: clause. For example, use
try:
import platform_specific_module
except ImportError:
platform_specific_module = NoneA bare except: clause will catch SystemExit and KeyboardInterrupt exceptions, making it harder to interrupt a program with Ctrl-C, and can disguise other problems. If you want to catch all exceptions that signal program errors, use except Exception:.
A good rule of thumb is to limit use of bare except: clauses to two cases:
- If the exception handler will be printing out or logging the traceback; at least the user will be aware that an error has occurred.
- If the code needs to do some cleanup work but then lets the exception propagate upward with raise; then try…finally is a better way to handle this case.
In addition, for all try/except clauses, limit the try clause to the absolute minimum amount of code necessary. Again, this avoids masking bugs. Yes:
try:
value = collection[key]
except KeyError:
return key_not_found(key)
else:
return handle_value(value)No:
try: # Too broad!
return handle_value(collection[key])
except KeyError: # <-- Will also catch KeyError raised by handle_value()
return key_not_found(key)Use string methods instead of the string module.
String methods are always much faster and share the same API with Unicode strings. Override this rule if backward compatibility with Python versions older than 2.0 is required.
Use ’‘.startswith() and’’.endswith() instead of string slicing to check for prefixes or suffixes. startswith() and endswith() are cleaner and less error-prone. Yes:
if foo.startswith('bar'):No:
if foo[:3] == 'bar':The exception is if your code must work with Python 1.5.2 (but let’s hope not!).
Object type comparisons should always use isinstance() instead of comparing types directly.
Yes:
if isinstance(obj, int):No:
if type(obj) is type(1):
When checking to see if an object is a string, keep in mind that it might be a Unicode string too. In Python 2.3, str and unicode have a common base class, basestring, so you can do the following:
if isinstance(obj, basestring):In Python 2.2, the types module has the StringTypes type defined for that purpose for example:
from types import StringTypes
if isinstance(obj, StringTypes):In Python 2.0 and 2.1, you should do the following:
from types import StringType, UnicodeType
if isinstance(obj, StringType) or \
isinstance(obj, UnicodeType) :For sequences (strings, lists, tuples), use the fact that empty sequences are false.
if not seq: if seq:No:
if len(seq) if not len(seq)Don’t write string literals that rely on significant trailing whitespace. Such trailing whitespace is visually indistinguishable, and some editors (or more recently, reindent .py) will trim them.
Don’t compare Boolean values to True or False using ==.
Yes:
if greeting:No:
if greeting == True:Worse:
if greeting is True:Regarding copyright, note that this document has been placed in the public domain.
A.4.3 Other guides for Python style
Although PEP 8 remains the most influential style guide for Python, you have other options. In general, these guides don’t contradict PEP 8, but they offer wider examples and fuller reasoning about how to make your code Pythonic. One good choice is The Elements of Python Style, freely available at https://mng.bz/dXlw. Another useful guide is The Hitchhiker’s Guide to Python, by Kenneth Reitz and Tanya Schlusser, also freely available at https://docs.python-guide.org.
As the language and programmers’ skills continue to evolve, there will certainly be other guides, and I encourage you to take advantage of new guides as they’re produced, but only after starting with PEP 8.
A.5 The Zen of Python
The following document is PEP 20, also referred to as “The Zen of Python,” a slightly tongue-in-cheek statement of the philosophy of Python. In addition to being included in the Python documentation, “The Zen of Python” is an Easter egg in the Python interpreter. Type import this at the interactive prompt to see it.
Longtime Pythoneer Tim Peters succinctly channels the BDFL’s (Benevolent Dictator for Life) guiding principles for Python’s design into 20 aphorisms, only 19 of which have been written down.
The Zen of Python
Beautiful is better than ugly.
Explicit is better than implicit.
Simple is better than complex.
Complex is better than complicated.
Flat is better than nested.
Sparse is better than dense.
Readability counts.
Special cases aren’t special enough to break the rules.
Although practicality beats purity.
Errors should never pass silently.
Unless explicitly silenced.
In the face of ambiguity, refuse the temptation to guess.
There should be one—and preferably only one—obvious way to do it.
Although that way may not be obvious at first unless you’re Dutch.
Now is better than never.
Although never is often better than right now.
If the implementation is hard to explain, it’s a bad idea.
If the implementation is easy to explain, it may be a good idea.
Namespaces are one honking great idea—let’s do more of those!
The Quick Python Book Fourth Edition
Naomi Ceder ● Foreword by Luciano Ramalho
System automation. High-performance web apps. Cloud and back-end services. Cutting edge AI. No matter what you’re building, it pays to know how to read and write Python! Th e Quick Python Book has helped over 100,000 developers get up to speed with the Python programming language. Th is revised Fourth Edition, fully updated for Python 3.13, explores the latest features and libraries and shows you how to code smarter with AI tools like ChatGPT.
The Quick Python Book, Fourth Edition teaches you the essential Python features and techniques you need for most common scripting, application programming, and data science tasks. Written for developers comfortable with another programming language, it dives right into the good stuff . New interactive notebooks, quick-check questions, and end-of-chapter labs all help practice and consolidate your new skills. Plus, you’ll fi nd practical advice on writing prompts and using AI assistants to accelerate your day-to-day work.
What’s Inside
- Python syntax, data structures, and best practices
- Object-oriented Python
- Must-know Python libraries
- Data handling
For beginning-intermediate programmers. No prior experience with Python required.
Naomi Ceder has been learning, teaching, and writing about Python since 2001. An elected fellow of the Python Software Foundation, Naomi is a past chair of its board of directors. In 2022 she became the seventh person to receive the PSF Distinguished Service Award.
For print book owners, all digital formats are free: https://www.manning.com/freebook
“Brilliantly crafted! It’s the book I recommend to smart people learning Python for the first time” —David Q Mertz, Service Employees International Union
“A good Python book should be on everybody’s bookshelf. Let me suggest The Quick Python Book!” —Delio D’Anna, cognitivplus
“I recommend it! It’s useful, clear, and engaging a combination that’s diffi cult to achieve!” —Doug Farrell, Author of The Well-Grounded Python Developer
“Hits the sweet spot between quick overview and deep dive.” —Trey Hunner, Python Morsels

